| RIKEN Center for
Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
| Chp1 and RNAi factors function in heterochromatin establishment | ||
September 27, 2004 - Chromosomes, with their distinct morphologies and well-known function as the storehouses of genomic DNA, are one of the most familiar structures in the biology of the cell. In eukaryotes, these organelles are manufactured from complexes of DNA, histones and other proteins, called chromatin. This complex organization spools and folds lengthy strands of genetic material into compact aggregates capable of fitting within the tiny space within the nucleus while fulfilling their function as depots and providers of information used by the cell's transcription machinery. The ability to condense itself so that genes needed for protein production remain accessible, while others remain knotted into the deeper recesses is central to chromatin organization, and is evidenced in the existence of two structurally different forms - a less condensed form called euchromatin and its more densely-packed counterpart, heterochromatin. In most contexts, heterochromatic regions do not permit the expression of any genes they might contain, owing to their extreme compactness and epigenetic modifications Heterochromatin is, nonetheless, important to the organization of non gene-encoding chromosomal regions necessary to the cell's ability to survive and replicate.
The chromosomes of the fission yeast, Schizosaccharomyces pombe , contain
a number of heterochromatic regions, including centromeres, telomeres and the
mating-type region. The configuration and function of chromatin in these domains
is studied as a model of how chromatin organization achieves its repressive
effects in other species, including our own. Generally, the formation of higher-order
chromatin structure can be divided, at least, into two processes; the establishment
and maintenance. Researchers at the RIKEN Center for Developmental Biology (CDB;
Kobe, Japan) Laboratory
for Chromatin Dynamics (Jun-ichi Nakayama, Team Leader) now report the identification of a role for the protein, Chp1, in the establishment
of heterochromatin. This protein had previously been implicated as important
to chromatin organization by studies that showed that yeast lacking the chp1 gene
suffered defects in chromosome segregation and centromeric transcriptional silencing,
and that the Chp1 localizes to centromeric heterochromatic regions. It is also
related to other known chromatin assembly molecules by virtue of its possession
of a conserved motif, known as the chromodomain, shared by many of the protein
players involved in epigenetic control of gene expression. Different chromodomain-containing
proteins have been thought to play discrete roles in the chromosome's various
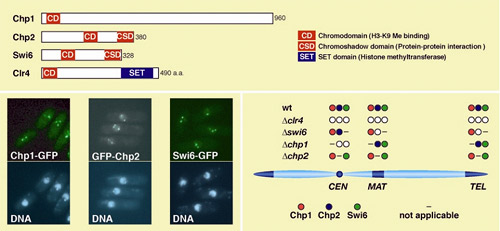
heterochromatic regions. Nakayama and colleagues started to analyze the localization of three
chromodomain proteins, Chp1, Chp2, and Swi6. Swi6 is a homolog of heterochromatin
protein 1 (HP1) in mammals, and has been shown to play a crucial role
in the formation of heterochromatin. The researchers found that Chp1 does
indeed associate with all three heterochromatic regions in S. pombe ,
the first demonstration of its presence outside of the centromere . A
detailed analysis of the differences in the localization patterns of Swi6
and Chp2 in mutant strains of fission yeast lacking Chp1 suggested that
this protein plays a vital role in the localization of Swi6 and Chp2 specifically
to the centromeric heterochromatin. This sparked the team's interest in whether its function might be linked
to RNA interference (RNAi), a nearly universal process responsible for
post-transcriptional gene silencing and known to be linked to both the
establishment of heterochromatin and to centromere-specific gene silencing
activity in fission yeast. Their further experiments showed that loss
of chp1 function had similar effects on the accumulation of
RNA transcripts to that of the loss of RNAi machinery components, indicating
a role in either the production or processing of centromeric RNA, either
of which might involve heterochromatin establishment. These similarities
suggest that RNAi machinery and Chp1 work together; however, it remained
unclear why mutations in chp1 or RNAi cause centromere-specific
defects. Given Chp1's universal association at centromeres, telomeres
and the mating-type region, they reasoned that the protein must have common
function in all heterochromatic domains. Histone proteins in chromatin, heterochromatin in particular, are subject
to a form of epigenetic modification called methylation, which affects
the expression of genes within the methylated region, generally by inactivating
them. The introduction of methyl modification on histones is thought to
be an initial and critical step in the establishment of heterochromatin.
Nakayama and colleagues designed experiments to analyze the establishment
steps using the histone methyltransferase, Clr4. When the function of
the gene, clr4 , is disturbed, methylation is drastically reduced;
reintroduction of clr4 restores this defect. However, in a chp1 mutant,
the restoration of clr4 failed to re-establish methylation not
only at centromeres, but also at the mating-type region and telomeres.
These experiments elegantly demonstrated the common function of Chp1 in
the establishment of all heterochromatic domains. Interestingly, tests
of the chromodomain proteins Swi6 and Chp2 revealed that both were essential,
possibly overlapping, factors in the maintenance of H3-K9 methylation.
They concluded that these three chromodomain proteins play distinct and
cooperative roles in the establishment and maintenance of heterochromatin
structure.
|
||
|
||
[ Contact ] Douglas Sipp : sipp@cdb.riken.jp TEL : +81-78-306-3043 RIKEN CDB, Office for Science Communications and International Affairs |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |