| RIKEN Center for
Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
| At the heart of the clock: Key genetic regulators that help the body keep time | ||
January 23, 2005 – Many of the mammalian body’s systems are on a tightly controlled schedule, their activity rising and falling in response to the natural cycle of darkness and daylight, but also in response to internal genetic regulators ticking away in specific sites of the brain and other organs. These “clock” genes work in a complex and interconnected system of regulatory loops in which temporal regulators of genetic activity themselves may be regulated by other repressors and activators. This intricate network represents fertile ground for study by systems biologists, who seek to identify and analyze higher-order organizations and coordinated behaviors in sets of interacting biological components. Scientists have been working for decades on elucidating the individual genetic components of these complex circadian systems in a range of experimental models. Now, in a study published in the online edition of Nature Genetics, Hiroki R. Ueda (Team Leader, Laboratory for Systems Biology) at the RIKEN Center for Developmental Biology (Kobe, Japan) reports the identification of what appears to be the central players of a network of genes in the mouse that regulates daily biological rhythms.
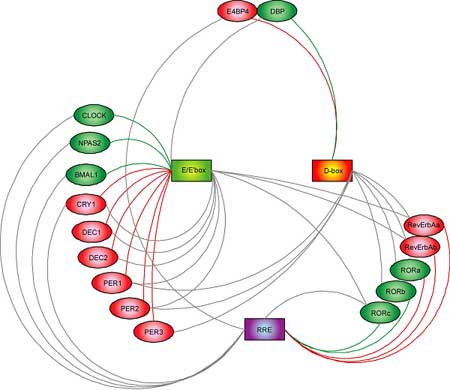
At least 16 clock and clock-related genes have been identified in the mouse, but to date the means by which they interact to achieve cyclical expression patterns has been a mystery. These 16 genes can be categorized into groups by their possession of one or more of three signature nucleotide sequences, conserved in human and mouse and linked to their times of peak expression. These three characteristic regions, named E/E′-box, D-box and RRE, are found on genes encoding transcription factors, which work to regulate the timing, site and amount of the production of RNA “messages” from gene sequences encoded in a cell’s DNA. The Ueda team found that each of the three classes of gene regulated transcription at different times of day. Specifically, “morning” E/E′-box regions were found on nine genes, D-box “midday” transcription factors were located on seven genes, and six genes regulated by “night” RRE domains were also identified. These three classes of transcriptions factors were also linked to the regulation of other genes in the 16-member network, with nine involved in morning transcriptional regulation, two in midday and five in night, allowing the team to devise a complex of model interacting regulatory loops in which individual genes both regulate and are regulated by other genes in the network. Analysis of the network showed that the E/E′-box morning transcription factors were the most prevalent of the three types, in terms of both the genes they acted directly on and the input they received from other genes in the system, with nine instances of each. By demonstrating that E/E′-box regulation is the most highly connected set of “nodes” within the network, the Ueda study provides compelling evidence that these factors are likely to play the most central role in circadian regulation and therefore may represent attractive targets for those interested in determining and analyzing means of controlling the network as a whole. This conjecture was borne out by at least one experiment, in which the overexpression of a repressor of E/E′-box regulation resulted in the disruption of the rhythmicity of normally circadian promoters, an effect not seen when repressors of D-box or RRE regulation were overexpressed. This detailed classification next allowed them to look for patterns in the activity of repressors and activators of clock gene expression, a search that revealed that different categories of genes tended to exhibit different temporal activity describable by two straightforward principles. If cycles of gene expression in this system are thought of as following the coordinated clockwise movement of hands around a dial, E/E′-box and RRE regulation could be characterized as “repressor precedes activator,” with the repressor gene always remaining slightly ahead of the activator as they cycle throughout the day. In D-box regulation, a separate principle applies, in which expression levels of repressors remain in near diametric opposition to those of activators, as if circling the dial in constant counterpoint. Although the exact means by which rhythmic oscillations in gene expression in human and mouse remain unknown, the “repressor precedes activator” mechanism appears to play an important role by delaying transcriptional activity, while “repressor antiphasic to activator” results in high-amplitude transcription. The Ueda study is an important first step toward the identification of systems governing circadian gene activity in the mouse, but much remains to be done before this ornately structured and tightly controlled network is fully understood, as molecular signaling cascades, chromatin modifications and the stability and localization of proteins are also thought to be involved in determining its cyclical rhythm. The benefits of such knowledge, however, could be great, as the ability to analyze and modulate normal and disturbed biological clocks in humans has potential clinical applications ranging from the development of drug administration regimens tailored to patients’ physiological rhythms to new approaches to the treatment of circadian disorders such as insomnia and depression.
|
||
|
||
[ Contact ] Douglas Sipp : sipp@cdb.riken.jp TEL : +81-78-306-3043 RIKEN CDB, Office for Science Communications and International Affairs |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |