| RIKEN Center for
Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
| Asymmetric T cell division by dint of Wnt | ||
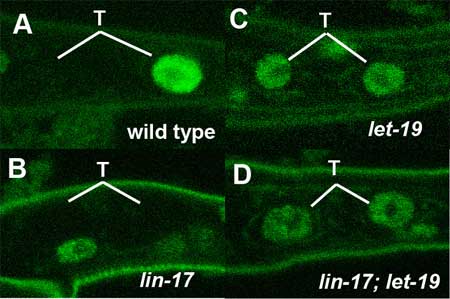
March 25, 2005 – Asymmetric division, in which a cell divides into daughter cells of different natures, is an indispensable process in the development of multicellular organisms. Every embryo begins its development as a single fertilized egg, dividing first to form a small cluster of very similar cells which later go on to differentiate into cells of myriad types, enabling the construction of complex tissues and organs. From the very first step of ontogeny, the ability to generate cells of different types from a single ancestor is central – indeed, without asymmetric cell division, the embryo would be mo more than an undifferentiated clump of identical cells. This asymmetry is achieved in different taxa by various means. The fruit fly Drosophila, for example, allocates the transcription factor Prospero to only one of two daughter cells during its neurogenesis, marking it as a ganglion mother cell directly responsible for generating neurons. The daughters deprived of Prospero, meanwhile, retain their identity as neuroblasts, progenitor cells capable of further self-renewal in subsequent rounds of cell division. Asymmetric cell division is important in many aspects of development in the nematode C. elegans as well, including the differentiation of a lineage of cells found in the tail region, named T cells. In a report published in the March 25 issue of the journal Development, Hitoshi Sawa and colleagues in the Laboratory for Cell Fate Decision, and in labs at Osaka, Kobe and Keio Universities, describe the requirement for a pair of molecules, LET-19 and DPY-22, thought to contribute to the formation of a multicomponent complex at work in T cell asymmetric division.
LET-19 and DPY-22 seem to achieve their asymmetry-regulating function by the transcriptional repression of target genes in the Wnt signaling cascade, a molecular pathway known to play roles many aspects of development in a diverse menagerie of organisms, including C. elegans. In T cells, for instance, the factor TLP-1 has recently been shown to be regulated by the Wnt pathway resulting in its unequal expression in T lineage daughters. Loss of function of the genes lin-44 or lin-17 (which respectively encode proteins homologous to the Wnt ligand and its receptor Frizzled) interferes with normal asymmetric division, causing the failure of T.p cells to generate the phasmid socket cells that would usually be included in their progeny. But the molecular means by which Wnt regulates the transcription of target genes in to achieve this asymmetry remains obscure. Sawa and colleagues first identified let-19 and dpy-22 in an analysis of worms in which the phasmid socket cells were absent. These phenotypes showed loss of asymmetry in T cell daughters similar to that seen in lin-17/frizzled mutants. Seeking to determine the mechanism by which let-19 and dpy-22 operate in this process, Sawa et al. looked at their relationship to a pair of genes, pop-1 and tlp-1, known to be expressed differentially in T.a and T.p cells, the anterior and posterior progeny of T cell division. In T.p cells, the localization of POP-1 is controlled by Wnt signaling, so the team used a construct fusing POP-1 with green fluorescent protein (GFP), allowing them to track the protein’s distribution within cells. These studies revealed that mutations of neither let-19 nor dpy-22 affected POP-1 localization, ruling out the possibility of a regulatory role for these genes in that process. They turned next to tlp-1, whose protein product is a downstream factor in the Wnt pathway expressed specifically in T.p but not in T.a cells. A study in lin-17 mutants showed the expected defects in tlp-1 expression. Looking at loss-of-function mutations for let-19 and dpy-22, Sawa found that in either of these mutants, tlp-1 expression could be seen in both the T.a and the T.p daughter cells, reflecting the loss of their normally unequal distribution. Worms in which both lin-17 and let-19 were mutated showed the same phenotype as the let-19 single mutant, indicating that let-19 works downstream of the Wnt signal. Further studies using other cell lineages failed to produce any effects on patterns of tlp-1 expression. Pursuing the let-19 and dpy-22 connection to the Wnt cascade, Sawa next examined their effects in Pn.p cells, a series of 11 cells found running down the anterior-posterior axis on the ventral (belly) side of the nematode. P1.p and P2.p, the anteriormost of the cells, and P9.p~P11.p, the last three in line, undergo fusion with the hypodermis, leaving the remaining six cells in the center to act as precursors of one of the worm’s organs of reproduction, the vulva. In this cohort, the gene bar-1 encodes the C. elegans homolog of β–catenin, which serves to maintain the expression of LIN-39/Hox, an inhibitor of cell fusion. In bar-1 mutants, fusion occurs ectopically, leading to a reduction in the number of vulval precursors. The team found that both the let-19 and dpy-22 mutants underwent less frequent cell fusion and seemed to repress the bar-1 phenotype characterized by ectopic fusion. Contrastingly, let-19 mutations did not suppress the lin-39 phenotype, suggesting that let-19 and dpy-22 work by repressing lin-39 downstream of bar-1. They looked for further involvement in other phenomena where Wnt signaling is a factor, such as endodermal induction and the formation of distal tip cells at the leading edge of the migrating gonad, but found that let-19 and dpy-22 function is specific to Pn.p and T cells. The gene products of let-19 and dpy-22 are homologous to mammalian MED13 and MED12, respectively, both of which are components of transcriptional regulatory “Mediator” complexes, which have been shown to play roles in the activation or repression of numerous genes. Two mammalian Mediator complexes – CRSP and ARC-L – are known, the latter being a larger, transcriptionally active complex that contains both MED12 and MED13. Co-immunoprecipitation assays demonstrated that LET-19 and DPY-22 bound other putative Mediator complex components, leading the team to surmise the existence of a pair of Mediator complexes in C. elegans as well, with LET-19 and DPY-22 acting as components of the counterpart to mammalian ARC-L. The results of this thoroughgoing investigation point clearly to roles for LET-19 and DPY-22 in both the asymmetric division of T cells and the regulation of Pn.p cell fusion by the inhibition of Wnt target genes. They speculate that activation of the Wnt (LIN-44/LIN-17) pathway results in the conversion of the ARC-L like complex to one that resembles CRSP by causing the release of LET-19 and DPY-22, which suppress the expression of tlp-1. Normal asymmetry may arise in T cells where the ARC-L-like complex inhibits tlp-1 in the anterior T.a cells, while the CRSP-like version lacking LET-19 and DPY-22 allows tlp-1 to be activated in the T.p posterior daughter. Similarly, in Pn.p cell fusion, LET-19 and DPY-22 may inhibit the expression of lin-39 in the absence of Wnt signaling, and conversely activating lin-39(and thereby repressing cell fusion) in the presence of Wnt signal. Although the means by which Wnt signaling and responsivity are regulated remain to be worked out, the Sawa study has shown how this prevalent and much-studied cascade may act as a kind of switch by adjusting the makeup of a protein complex responsible for regulating gene transcription. |
||
|
||
[ Contact ] Douglas Sipp : sipp@cdb.riken.jp TEL : +81-78-306-3043 RIKEN CDB, Office for Science Communications and International Affairs |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |