| Standing the Test of Time: Transcriptional Feedback in the Mammalian Clock |
 |
 |

February 13, 2006 – Researchers studying the cyanobacterium, Synechococcus elongatus, gave the circadian biology community something of an unexpected wakeup call last year when they published their findings on how this primitive organism keeps time. Most classical models intended to describe the activity of biological clocks suggest that the autoregulatory mechanism relies on transcriptional negative feedback. In a simplified version of this clockwork, one set of genes acts to inhibit the transcription or translation of a second set, whose function is, somewhat paradoxically, to activate the first; this is mechanism believed to be universal to all circadian biology. The S. elongatus study, however, showed otherwise, for the molecular clocks in these unicellular creatures happily kept ticking away even when transcriptional regulation was factored out. This single report prompted a re-examination of what had been up to that point an article of faith, in which scientists found themselves scrambling to substantiate that feedback repression is indeed one of the mainsprings of the circadian clock in other species.
The hunt for direct evidence has now paid off, with the results of a thoroughgoing joint study by members of the Laboratory for Systems Biology (Hiroki R. Ueda; Team Leader) and colleagues in several American labs, including the Scripps Research Institute, which conclusively demonstrated the requirement for transcriptional feedback repression in the mouse biological clock function. The work, which involved the meticulous dissection of the murine timekeeping network, showed that CRY and PER proteins regulate their own expression by repressing the formation of a functional complex by the transcriptional activators CLOCK and BMAL1, which themselves were suspected to form a complex that binds to promoters of CRY and PER.
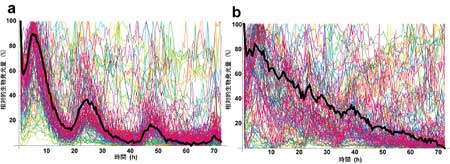 |
| Circadian rhythm is lost in cells carrying mutations in biological clock genes. |
To verify the feedback repressor model, the collaborators first identified mutations in the CLOCK and BMAL1 genes that showed resistance to inhibition by the Cry-encoded protein, CRY1. This desensitization was traced to defects in a pair of regions, one in the CLOCK PAS domain, the other in the C-terminus of BMAL1. Interestingly, mutation of both genes together showed a combinatorial effect; the double mutants were much less responsive to repression by CRY1 than either of the mutations on its own. Subsequent biochemical experiments indicated that CRY1 is be less able to bind to CLOCK/BMAL1 complexes in which either or both of the constituent proteins contained a mutation. Additional findings involving the other putative set of transcriptional repressors, the PER proteins, tended to confirm that physical interaction between CRY and PER and the CLOCK/BMAL1 complex is responsible for the normal inhibitory effect, and that this interaction is interfered with in mutant CLOCK/BMAL1 complexes. Such mutations had a crossover effect as well, as CRY-insensitive CLOCK/BMAL1 phenotypes showed a loss of circadian rhythmicity in Per2 and Bmal1 promoter activity. Population- and single-cell-level monitoring of the cellular rhythms of mammalian cells carrying the mutations in question reinforced the importance of CRY-mediated repression; at both scales, normal circadian patterns were disrupted.
This latest work from the Ueda lab bolsters the old case for the centrality of negative feedback in the form of a transcriptional repressor in the regulation of the mammalian cellular rhythms. Of course, it is likely that there are additional components to this intricate clockwork that continue to cycle even when transcriptional feedback is stopped (as is the case in S. elongatus), but we know now that these alone are insufficient to keep our cells running on schedule. Perhaps more importantly, an improved understanding of the body’s molecular clock might some day lead to therapies for individuals whose daily rhythms are out of sync. “If we could find a way to control these circadian regulatory mechanisms,” Ueda notes, “it might open up new treatments for common medical problems such as sleep disorders, which could stand as a real benefit for people on non-standard wake-and-sleep schedules such as those working irregular shifts.”
|

