| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
The demand for large quantities of faithfully amplified mRNAs from a single cell have now become fulfillable, thanks to a virtuoso set of refinements to the polymerase chain reaction (PCR) method developed by Mitinori Saitou (Team Leader, Laboratory for Mammalian Germ Cell Biology; Researcher, PRESTO program, Japan Science and Technology Agency), Kazuki Kurimoto, Yukihiro Yabuta (Laboratory for Mammalian Germ Cell Biology) and colleagues in the CDB. And, in a proof-of-principle demonstration published in the same March 17 article in Nucleic Acids Research, the authors show that the technique can be used to characterize individual cells from the inner cell mass of the blastocyst, revealing that this developmentally important population actually comprises two distinct groups of cells.
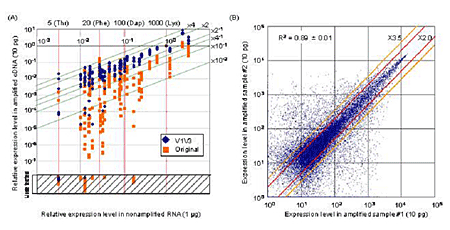
The high-density oligonucleotide microarray analyses performed in this research involve a number of preparatory steps, first of which is the high-fidelity amplification of cDNAs complementary to the full set of messenger RNAs transcribed by a cell. Of the extant methods for amplification, PCR, with its exponential yields, is the most suited to the tiny amounts of mRNA obtainable from single cells, but a previously reported single-cell PCR method tends to produce by-products as the result of the non-specific amplification of misused primer-derivatives, has been dogged by problems of bias and misrepresentation, and often fails to yield mRNAs of reproducible quality. Here, as with so many other amplification strategies, the question of how to boost signal without doing the same for noise remains fundamental. Kurimoto et al’s achievement was to overcome the shortcomings of PCR without sacrificing its amplificatory power, by a wedding of attributes of both exponential and linear strategies. The team first obtained RNAs expressed in embryonic stem cells and diluted them into a single-cell level. They then made quantitative comparisons of the pre- and post-amplification levels of 23 genes. They found that by eliminating unreacted primers at an early stage, and by limiting the number of cycles of amplification in the first round to 20, they were able to reduce distortion and the generation of by-products dramatically. These analyses also identified a pair of primers, V1 and V3, that, when used in combination, produced much greater efficiencies over single-primer schemes by allowing the retention of directionality of cDNA amplification products. This two-primer strategy now makes it possible to linearly amplify the complementary RNAs used in microarray experiments from a single strand of DNA. These, and the team’s many other refinements have resulted in a method of PCR that provides better representation, reproducibility and coverage than ever before, with single-cell accuracies of 97% true-positive detection of transcripts present as 20 or more copies per cell, and 93% for those expressed at levels of as low as 5 copies. Eager to prove the utility of the new strategy, Kurimoto et al. performed microarray analyses of individual cells from the inner cell mass of day 3.5 mouse embryos. Even this first proof-of-concept experiment yielded new insights, when the authors found that the inner cell mass, which was thought to be homogeneous at this stage of development, actually comprises cells expressing genes characteristic of a pair of more differentiated populations, primitive endoderm (PE) and epiblast. This represents the first demonstration that the morphological segregation of PE and epiblast at day 4.5 may in fact have its underpinnings in changes in gene expression a full day earlier in embryogenesis. By enabling the analysis of genome wide expression patterns at the single-cell level, the methodological advancements described in the Saitou report are poised to stand as a watershed in the study of a broad range of biological questions. Thanks to this technology, early developmental processes and the mechanisms of fate determination in somatic stem cell systems, phenomena which have long eluded explication, now await study at single-cell resolution. |
|||||
|
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |