| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
Now, in a study by Shinsuke Aramaki and Kohei Hatta of the Laboratory for Vertebrate Body Plan (Shinichi Aizawa; Group Director) the use of such visualization has been elevated to a new plane through the use of Dronpa, a GFP with a unique and striking photosensitivity. On excitation by wavelengths of light of about 490 nm, the Dronpa protein undergoes photobleaching and ceases to glow; but remarkably, its powerful fluorescence is restored following irradiation at around 400 nm. In this study, published in Developmental Dynamics, Aramaki and Hatta took advantage of this novel set of photochemical properties for the first time in visualizing cellular-level structures in living zebrafish embryos.
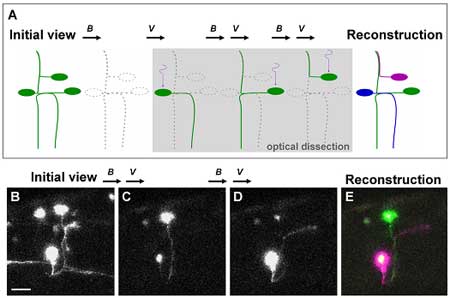
After establishing the feasibility of using Dronpa in zebrafish embryos the authors, through a series of novel techniques, were also able to analyze the anatomy of neural networks. Perhaps most importantly, they succeeded in visualizing a number of overlapping individual neurons that had previously proved difficult to view in isolation. Focusing on neurons found in complex wiring patterns they dissected neural networks to the single cell level following injection of a UAS-dronpa construct. In these transgenic fish, irradiation followed by a reactivation of the green fluorescence highlighted the entire morphology of each neuron as the protein diffused into the neurites. A subsequent merger of these isolated neuronal images enabled the reconstruction of neural networks in which single neurons were distinguishable by the different pseudo-colors used to label them. Further application of the technique included an attempt to locate the position of neuronal somata from visible axons by retrograde labeling, in which Dronpa was first eliminated from the area of interest by irradiation, followed by a further irradiation of the axon of interest. On attempting to obtain multiple bright images, however, Aramaki and Hatta found that the relatively high intensity required to eliminate Dronpa at 488 nm often resulted in the attenuation of signal. In an attempt to overcome this they decided to irradiate neurons of interest each time, or after every few times, to illuminate it to a sufficient brightness when scanning for observation. This ultimately resulted in images with considerably stronger fluorescence. The authors also used two-photon confocal microscopy, which made it possible for Dronpa to be activated by a longer wavelength of laser light to illuminate cells buried deep in tissue while avoiding activating neighboring cells in the light path. Hatta’s interest in Dronpa was first piqued when a fellow CDB researcher suggested it to him following a presentation he gave in 2004. “It wasn’t easy to adjust the conditions to allow us to switch the fluorescence on and off in vivo”, he admits, “so I was very pleasantly surprised to see that it finally worked so well in zebrafish embryos.” The success of Aramaki and Hatta’s use of Dronpa has large-scale and high-throughput potential, as the repetitive cycle of photobleaching, reactivation and visualization lends itself to automation.The non-invasive highlighting of specific structures and organs by Dronpa will also assist in their dissection or sorting for use in transplantation or gene-expression experiments. A forthcoming study by Hatta also points to uses for a similar technique in mammals, offering all the advantages of this novel system in a broader range of biological models. |
|||||
|
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |