| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
Now, in a study published on April 30 in the advance online edition of Nature Cell Biology, Yasushi Izumi and colleagues working in the Laboratory for Cell Asymmetry (Fumio Matsuzaki, Group Director) report that the Mud (mushroom body defect) protein coordinates the orientation of the mitotic spindle with cell polarity during asymmetric division of neuroblasts in the fruitfly, Drosophila. “This is the first time anyone has been able to show a general mechanism behind the coupling of cell polarity and axis of division,” notes Matsuzaki, “which is exciting, as it may help us develop new insights into the means by which cells switch between proliferative and differentiative modes of division.” These findings, which were made with support from the Japan Science and Technology Corporation CREST program and in collaboration with the Institute for Medical radiation and Cell Research at the University of Wuerzburg (Germany), are of particular interest due to the highly evolutionarily conserved nature of the Mud protein, which has homologs in both C. elegans and vertebrates, all of which appear to be involved in regulating spindle orientation vis-à-vis the cell cortex.
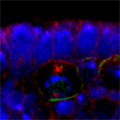
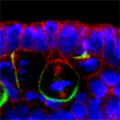
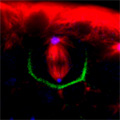
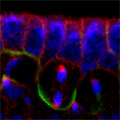
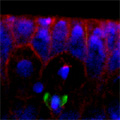
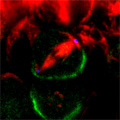
Looking at Mud’s intracellular distribution, they found that it localized to the apical side of neuroblasts throughout the cell cycle as well as their centrosomes (the central hub from which spindle microtubules radiate) during the mitotic phase, in contrast to its binding partner Pins, which showed up only at the apical cortex. Similarly, in symmetrically dividing epithelial cells, both Mud and Pins were found at the lateral cortex while Mud alone appeared at the centrosomes. In both pins and Gαi loss-of-function mutants, Mud failed to be recruited to the apical cortex of mitotic neuroblasts, but interestingly, during interphase, when the cells are not dividing, Mud localization was unaffected in pins mutant cells. Contrastingly, centrosomal Mud accumulation was found to rely on microtubules, rather than the apical complex, suggesting that Mud distribution is governed by a pair of independent systems: recruitment to the apical cortex by the Pins-Gαi complex and to the centrosome via microtubules. Importantly, the Mud-Pins interaction works independent of the mechanism determining the localization of cell fate determinants, such as Prospero and Miranda, that function to ensure the asymmetry of neuroblast division; epithelial cells, which divide symmetrically and perpendicular to the apical-basal axis, also rely on this association to orient their division axes toward cortically localized Pins. The question then was, how does Mud function in aligning the mitotic spindle along the apical-basal axis and thus enable the asymmetric parceling of cell fate determinants so crucial to fly neurodifferentiation? Mud mutant neuroblasts retain their cortical polarity, but the orientation of their mitotic spindles is severely compromised. This failure of “spindle coupling” is similar to a defect noted in Pins mutants, but with two significant differences: first, in Pins mutants, both the cortical factors and the spindle misorient, while only the spindle is askew in Mud-deficient flies; and second, spindle coupling recovers during telophase in pins, but not mud, mutants. These differences in phenotype point to incompleteoverlap of function between Mud and the Pins-Gαi complex, resulting in distinct and independent roles at different mitotic stages. In addition to their unequal inheritance of cell fate determinants, size matters in asymmetric daughters as well. Watching Mud mutant neuroblasts divide, Izumi and colleagues observed that the angle of spindle orientation appeared to be linked to daughter cell size asymmetry, with highly skewed spindles tending to give rise to daughter cells closer to each other in size (which is abnormal in neuroblast division). In the most extreme cases, two daughter cells of equal size were produced. Spindle uncoupling in these mutants was also frequently accompanied by the formation of additional centrosomes, indicating the Mud may work in the establishment and/or maintenance of these structures. This set of findings suggests a new model for spindle coupling, in which Pins is both necessary and sufficient to recruit Mud to the cortex during mitosis, and that the location of this Pins-Mud complex determines spindle orientation, enabling cells to halve as they should. Tantalizing evidence already exists that other taxa share this molecular mechanism – in vertebrates, NuMA associates with the Pins homolog LGN to regulate spindle movement, while in the nematode, Lin-5 complexes with GoLoco proteins and Gα to generate the pulling force exerted by astral microtubules in the lead-up to the first embryonic cell division. The exact manner in which Mud interacts with the cytoskeleton during mitosis and its function in centrosome organization await further study; for now, this work by the Matsuzaki group shed new light on the extraordinary precision and coordination of processes that tell cells how and where to cleave apart. |
|||||||||||||||||||||||
|
|||||||||||||||||||||||
|
|||||||||||||||||||||||
 |
|||||||||||||||||||||||
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |