| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
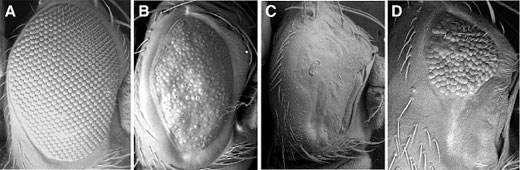
Previous work by Tsuda and colleagues had revealed that a protein complex that includes Ebi, SMRTER, Su(H) (Suppressor of Hairless) and strawberry notch, serves to upregulate the expression of the intercellular signaling factor, Delta (Dl), an intriguing result, given that the Ebi/SMRTER/Su(H) complex was known to function generally as a repressor, rather than an activator. On conducting a screen for genes repressed by Ebi/SMRTER/Su(H), the group found that a gene named charlatan was downregulated by the finely tuned interaction between Su(H) and Ebi, a corepressor whose activity had been shown in earlier work to rely on SMRTER. Using Gene Search technology, in which genes can be detected by monitoring phenotypic changes caused by GAL4-dependent misexpression of regions of DNA adjacent to vector inserts, they were able to narrow the site of activity of the Ebi-dependent transcriptional repression to the vicinity of the charlatan promoter, PR(chn). Chromatin immunoprecipitation (ChIP) next confirmed that both Ebi and its corepressor SMRTER associate with the charlatan promoter region in vivo. Turning to the third element of the signaling complex, Su(H), Tsuda examined how it might interact with PR(chn). Su(H) is a critical downstream factor in Notch signaling, in which binding of a ligand (such as Delta) to Notch results in the cleavage of the receptor molecule and the release of the intracellular domain NICD into the cytoplasm, where it makes its way to the nucleus, forms a complex with Su(H) and activates gene transcription. Additional ChIP analyses showed that the activator, NICD, and the Ebi-SMRTER corepressors are targeted to the same binding site on the charlatan promoter, such that binding by one prevents binding by the other. Intrigued by this shared binding site, the Hayashi group searched for genetic homologs of charlatan and identified a range of structural and functional similarities between Drosophila chn and the human gene NRSF/REST. Given the role of NRSF/REST in human neural regulation, Tsuda et al. looked for analogous function in Drosophila and found that a consensus binding sequence for Chn (which they named CBE, for Charlatan-binding element) was present in a number of neural genes for which the human homologs are targeted by NRSF/REST. With this knowledge in hand, they found that Delta (a Notch ligand) includes two CBEs, suggesting that it is a target of Charlatan. Testing this, they found that ectopic expression of chn in committed photoreceptor cells reduced Delta levels, blocking cone cell differentiation without affecting their basic neuronal status. The sum of these findings points to a scenario in which Ebi and Charlatan, both of which are repressors, have a net positive effect on the regulation of Delta in photoreceptor cells. In this “double-negative” model, ebi represses chn to permit expression of Delta, which then induces cone cell differentiation. Charlatan’s role seems to be restricted to early stages of eye development, as, once the process has been initiated, the requirement for chn fades. “It was very pleasant surprise for us to identify a fly homolog of human NRSF/REST when looking for downstream targets of another evolutionarily conserved gene, Ebi/TBL1,” admits Hayashi. “We’re hopeful that these findings will lead to a clearer understanding of the crosstalk between EGFR and Notch signaling in neural differentiation.” |
|||||||||
|
|||||||||
|
|||||||||
 |
|||||||||
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |