| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
Now, work by Tetsushi Iida of the Laboratory for Chromatin Dynamics (Jun-Ichi Nakayama; Team Leader), shows that, in the fission yeast Schizosaccharomyces pombe, the ribonuclease Eri1 breaks down small interference RNAs derived from heterochromatic regions, thereby controlling heterochromatin assembly. These findings are of particular interest, as the molecular players involved are widely conserved across the biological spectrum, including in man. The results of the Nakayama team’s study were published in the June 22 online edition of Current Biology.
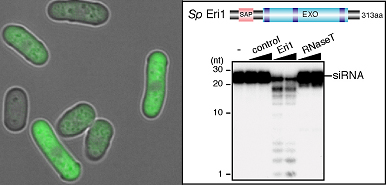
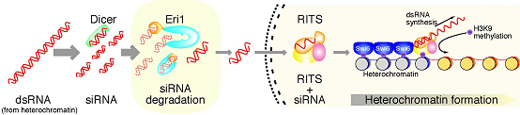
Iida et al began by identifying the S. pombe ortholog of Eri1, and verifying that it too had ribonuclease activity against siRNAs in vitro. Zeroing in on a possible role in heterochromatin formation, they engineered mutant yeast cells that lacked eri1, and found that centromeric and other heterochromatic regional gene expression was repressed, apparently due to the absence of Eri1 enzymatic activity. They next showed that Eri1’s function is linked to the RNAi machinery, which was already known to control the heterochromaticity of these chromosomal precincts. From this evidence, they surmised that Eri1 has a negative regulatory effect on heterochromatic gene silencing, by targeting and breaking down specific siRNAs. In S. pombe, heterochromatin is defined by the highly specific protein modifications (the methylation of histone H3 at lysine 9), which, thus marked, becomes a target for the factor Swi6, leading to the tight winding for the affected sequence into a gene-silencing heterochromatic coil. In eri1 mutants, histone methylation and Swi6 were enriched, and heterochromatin assembly at the centromeres upregulated. These same mutants also saw an increase in the abundance of siRNAs generated by the RNAi machinery, as evidenced by their increased association with the RITS complex, which suggests that eri1 normally functions to downregulate the buildup of siRNAs and their binding to RITS complexes.
The findings from these experiments led the Nakayama team to develop a model of heterochromatin assembly in fission yeast in which Eri1 enzymatically attacks heterochromatic region-derived siRNAs which would otherwise activate the RITS complex to drive further heterochromatin assembly, creating a perfectly titrated set of molecular checks and balances that keeps the cell’s gene expression in tune. “It looks like Eri1 pulls off the neat trick of ensuring that heterochromatin assembles only where it should by maintaining siRNAs at just the right level,” says Nakayama. “We’ve already begun to see from work in other labs how Eri1 can affect siRNA accumulation in C. elegans under certain conditions, so it will be exciting to find out more about its range of functions in RNAi.” |
|||||||||||||||
|
|||||||||||||||
 |
|||||||||||||||
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |