| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
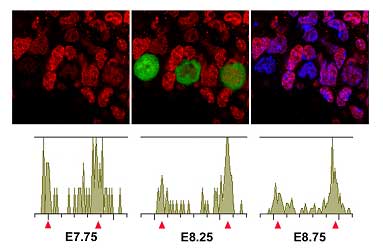
The team began by comparing chromatin modifications in PGC precursors (which express the germ cell specifying gene Blimp1) with somatic neighbors and found that there were still no significant differences in epigenetic marking at this early stage before the PGCs begin their migration. Immunohistochemistry using antibodies against histone modifications performed slightly later (starting at about embryonic day 7.75), however, showed that H3K9me2 levels progressively declined in PGCs, dropping to very low levels by E8.75. The team speculates that this may be due to the downregulation of the histone methyltransferase GLP in migrating PGCs. A second histone modification, H3K27me3, conversely became upregulated, starting about a half-day later at E8.25 and continuing until E9.5, at which point about 85% of the cells stained strongly for H3K27 trimethylation. Methylation of histones frequently represses the transcriptional activity of the genes included in the affected chromatin region, so Seki et al. next asked whether transcription in PGCs is affected by this massive reprogramming. When they looked at levels of RNA polymerase II (RNAP II) phosphorylation, which serves as a good index of global transcriptional activity, they were surprised to find only low levels of this modification. The repression of RNAP II-dependent transcription appears to begin in migrating PGCs at about E8.0 and to occur by a yet-unknown mechanism that interestingly does not rely on chromatin-based silencing. Although questions about the mechanistic details remain, the Saitou team now have a timeline of the events in PGC reprogramming, in which H3K9me2 first starts to decline, followed by the transient loss of RNAP II phosphorylation, the upregulation of H3K27me3, and finally the resumption of RNAP II-dependent transcription. Noting that the PGC population appears to undergo a slowdown in cell proliferation between E8.0 and about E9.25, the team next turned to analyze the mitotic activity of these cells. BrdU labeling showed that the majority of PGCs during this period failed to enter the S-phase of mitosis. Expression levels of cyclin B1, which accumulates in the cytoplasm of G2-phase cells, suggested that PGCs are arrested at the G2 phase of the cell cycle during their migration. They finally confirmed this by directly measuring the DNA contents of migrating PGCs by FACS analysis, shoring up the case that the epigenetic reprogramming most likely occurs in the G2-phase of the cell cycle. “The mammalian germline system may be the best in vivo system with which one can study the precise mechanisms of genome-wide epigenetic reprogramming,” says Saitou, “since germ cells continuously reprogram their epigenome by genetically tractable pathways throughout their development.We hope that these studies will provide essential insights into the reprogramming of somatic cell nuclei in general.” |
|||||
|
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |