| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
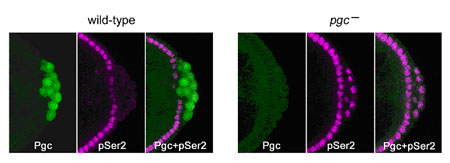
Kazuko Hanyu-Nakamura, Hiroko Sonobe-Nojima and colleagues in the Laboratory for Germline Development (Akira Nakamura; Team Leader) have now identified the factor responsible for such transcriptional repression in Drosophila germ cell precursors. Their work, published in Nature, reveals that the product of the polar granule component (pgc) gene represses Ser2 phosphorylation of the RNAPII carboxy-terminal domain (CTD) in germline progenitors known as pole cells by interfering with the recruitment of a second factor, P-TEFb, which is known to play a role in CTD Ser2 phosphorylation in vivo. The project began somewhat serendipitously when the Nakamura team noted that pgc RNA, which was assumed to be non-coding, in fact contains a sequence that suggests a translation start site. Going back to the completed Drosophila genome, they found that pgc should indeed be capable of encoding a small protein, which they verified by immunostaining; antibodies against the Pgc peptide showed immunoreactivity in early-stage pole cells, before tapering off later in embryonic development. They next generated flies with various mutations causing losses of pgc-function and discovered that, while pole cells did form, there was no repression of Ser2 phosphorylation in the RNAPII carboxy-terminal domain, causing the cells to degenerate by mid-stage embryogenesis. This defect could be rescued by the expression of intact Pgc. Moreover, misexpression of pgc in somatic cells resulted in the repression of Ser2 phosphorylation, strongly suggesting that the newfound protein plays a central role in transcriptional repression. The team next turned their attention to a second factor, P-TEFb (Positive transcription elongation factor b), which had previously been implicated in the phosphorylation of CTD Ser2 and promotion of productive transcription elongation. Suspecting that this might be a target of Pgc, they used co-immunoprecipitation and pull-down assay to establish a specific interaction between the two factors, both in vitro and in vivo, in which Pgc interacts with Cdk9, a catalytic subunit of P-TEFb. Overexpression of P-TEFb in pole cells produced a similar effect to that of loss of pgc function, suggesting that Pgc represses Ser2 phosphorylation in pole cells by interfering with the function of P-TEFb. But the finding that an MBP-Pgc construct, which interacted with P-TEFb, did not affect CTD phosphorylation by that factor in vitro, indicated that Pgc’s function was unlikely to regulate the catalytic activity of P-TEFb. They compared localization of P-TEFb in untreated somatic tissue and tissue in which Pgc was misexpressed, and found a dramatic reduction of their normal localization to active promoter regions on chromosomes. This suggested that Pgc may function by sequestering P-TEFb and preventing its recruitment to promoter sites, a view that was shored up by the finding that Pgc expression inhibited normal P-TEFb recruitment to heat shock genes following heat shock. “Studies in C. elegans have shown that another germline protein, PIE-1, is also involved in regulating the phosphorylation of CTD Ser2 and that it appears to bind to the CyclinT subunit of P-TEFb,” says Hanyu-Nakamura. “What’s interesting is that Pgc and PIE-1 are non-homologous, which means these germline transcriptional repression systems may well have evolved independently.” |
|||||
|
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |