| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
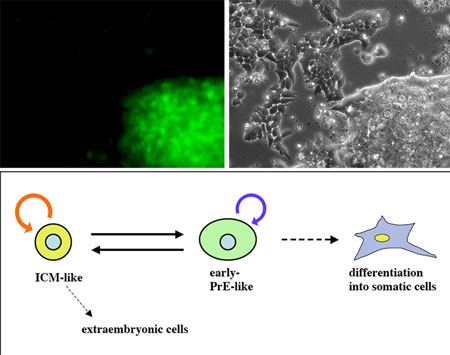
A new study by Yayoi Toyooka and colleagues in the Laboratory for Pluripotent Cell Studies (Hitoshi Niwa; Team Leader) comes down clearly in favor of the heterogeneity theory, showing that subsets of ES cells exhibit gene expression variations corresponding to the ICM, epiblast and primitive ectoderm stages of development. In a report published in Development, Toyooka, now at the University of Oxford, reveals that while all ES cells express the pluripotency marker Oct3/4, they show fluctuations in the expression of another gene previously taken as an ES hallmark, Rex1. The study began with the establishment of a system for visualizing the expression of these two genes using a pair of fluorescent proteins. When they cultured the cells using a method to select for Oct3/4-expressing cells, they found that some of these expressed Rex1 while others did not, with predominantly Rex1+ colonies showing compacted morphologies, and Rex1- colonies tending to be flatter. They used quantitative PCR to examine gene expression in the two subpopulations in more detail, and found that the overall expression patterns in Rex1+cells was similar to that seen in inner mass cells, while Rex1-cells expressed genes in patterns reminiscent of primitive ectoderm. Interestingly, when they separated these fractions into purified populations, they found that each was able to give rise to the other spontaneously; that is, Rex1+ cells showed up in Rex1- colonies, and vice versa. This worked even in colonies derived from a single Rex1-positive or negative cell. It has long been known that cells from the inner cell mass or epiblast can contribute to chimera formation when injected into blastocyst-stage embryos, but cells taken from primitive ectoderm cannot. Investigating whether this might be true of ES cells in which Rex1 expression is switched on or off, Toyooka et al. injected either Rex1+ or Rex1- cells into mouse blastocysts and found that, similar to ICM and epiblast cells, the Rex1+ cells contributed strongly to chimeric embryonic tissues, but that, as is the case for primitive ectoderm, the Rex1-cells did not. Given this apparent correspondence of Rex1+ and Rex1- cells respectively to the inner cell mass and primitive ectoderm stage of development, the Niwa team next checked whether their differentiation would follow a similar pattern. When differentiation was induced in colonies enriched for Rex1- cells by the withdrawal of LIF (a factor that keeps mouse ES cells in an undifferentiated state) they showed a lower tendency to give rise to extraembryonic tissue than did a control group, indicating another similarity with primitive ectoderm cells in vivo. Conversely, Rex1- cells were comparatively easier to steer down somatic pathways, such as mesoderm or neuroectoderm, than were their Rex1+ counterparts. “I suppose that many people who have worked with ES cells already suspected that they were not entirely homogeneous,” says Toyooka, “but now, by looking at Rex1 expression, we’ve been able to show that there are definite subpopulations in terms of both gene expression and differentiative potential. Perhaps this work will serve as a model for studying the control of fluctuations in gene expression.” |
|||||
|
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |