| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
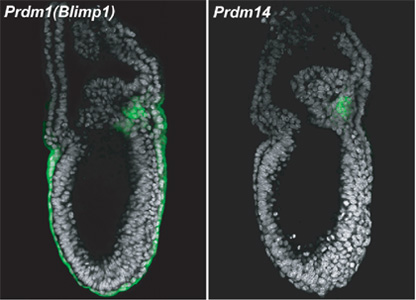
July 15, 2008 – Germ cells diverge from their somatic counterparts fairly early during mammalian development, undergoing at least three processes: the repression of somatic genes, the reacquisition of the potential for pluripotency, and subsequent epigenetic reprogramming to a committed germline fate. The genetic factors involved in germline specification have been traced as far back as day 6.25 of embryonic development, when the gene Prdm1 (also known as Blimp1) is switched on in a handful of cells in the epiblast, in what is believed to be the first critical step in the pathway to determining germline fate. A recent genome-wide study of transcriptional dynamics in early germline progenitors by the Laboratory for Mammalian Germ Cell Biology (Mitinori Saitou; Team Leader) has revealed, however, that the network is more diverse than previously expected, with Prdm1 acting as a sort of conductor keeping this genetic orchestra in harmony.
Now, Masashi Yamaji and others from the Saitou lab have discovered that a gene identified in their previous analysis, Prdm14, plays a critical role in the establishment of the germ cell lineage. In a study published in Nature Genetics, they report that this gene, a transcription factor expressed only in the germline, is necessary for two of the three hallmark events in the acquisition of germ cell fate. Prdm14 is first expressed transiently in the inner cell mass, a cluster of cells in the interior of the 3.5 day-old embryo that give rise to the embryo proper, but fades out by day 5.5, before reappearing at around E6.5 at a stage called the early streak, in which the germline master gene Prdm1 is also just beginning to be expressed. This early germline-specific expression prompted Yamaji et al. to look more closely for a role in the specification of primordial germ cells (PGCs). To do so, they established mutant mouse lines in which Prdm14 was deleted. The homozygous Prdm14-/- mutants were of normal appearance, but both the males and females were sterile, their gonads being entirely devoid of germ cells. The team examined germline development in the knockout embryos to determine the stage at which Prdm14 functions, and found that while somatic genetic programs (as represented by Hoxb1) were repressed, the cells failed to regain pluripotency (indicated by expression of Sox2) or to undergo the widespread epigenetic reprogramming indicated by the erasure of histone H3 lysine(K)9 di-methylation and upregulation of H3K27tri-metylation). Further in vitro experiments using primordial germ cells from the Prdm14 mutants showed that, as suggested by their inability to reinitiate Sox2 expression, they were also unable to de-differentiate to pluripotent embryonic germ cell-like cultures, which is another feature of wildtype PGCs. Interestingly, when Yamaji and colleagues analyzed the position of Prdm14 in the genetic network associated with PGC specification they found that although its initial activation does not require Prdm1, its subsequent maintenance and/or upregulation do. The trigger for Prdm14’s activation appears rather to be dependent on the Bmp4-Smad pathway, a separate signaling routine at work in the early mammalian embryo. “We now know that the specification of the germ cell lineage is orchestrated by the two transcriptional regulators, Prdm1 and Prdm14,” says Saitou. “Our next challenges will be to clarify the biochemical mechanism by which these two key factors function and to reconstruct germ cell specification in vitro. We hope that this work will provide a useful foundation for reproductive and regenerative medicine.”
|
|||||
|
|||||
|
|
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |