| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
September 18, 2008 – Mammalian brains feature dozens of neuronal subytypes generated principally by the division and differentiation of progenitor cells during embryonic development. In the neocortex, for example, elongated progenitor cells straddle and shuttle in the ventricular zone along this apical-basal axis, undergoing mitosis at the apical surface to give rise to the multiple strata of cortical neurons. But while molecular markers have been identified that distinguish apical progenitors from their basal counterparts, the mechanism that underlies their differentiation has remained largely a mystery. A thoroughgoing statistical analysis of gene expression profiles of individual neocortical progenitor cells by Ayano Kawaguchi and colleagues in the Laboratory for Cell Asymmetry (Fumio Matsuzaki; Group Director), as well as the Laboratories for Mammalian Germ Cell Biology (Mitinori Saitou; Team Leader) and Systems Biology (Hiroki R. Ueda; Team Leader), has now yielded new insights into the molecular basis of their heterogeneity. Using a combination of single-cell cDNA amplification, cluster analysis, and in situ hybridization, the group identified a set of genes that distinguishes the stem cell-like apical progenitors from their more differentiated basal counterparts, as well as a possible role for Notch signaling in maintaining these two progenitor populations. Their findings were published in the journal Development.
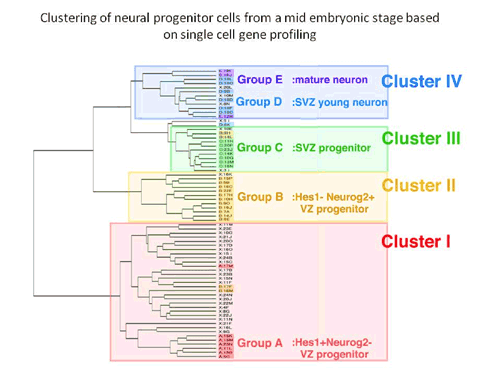
Kawaguchi began by manually selecting a random assortment of individual cells (including several mature neurons used as controls) from the ventricular/subventricular zone of embryonic mouse forebrains, and used a high-efficiency single-cell PCR technique to analyze their expression of a set of 20 marker genes known to be associated with various populations of progenitors and neurons. This profiling work enabled them to class the cells into five groups: undifferentiated and neuronally-biased apical progenitors from the ventricular zone, subventricular (basal) progenitors, young neurons, and mature neurons. To validate these initial findings, the group next performed hierarchical cluster analyses of the cells using microarray data. In both supervised and unsupervised analyses, the gene expression profiles of the individual cells allowed them to be grouped into four clusters closely corresponding to the results of the cDNA analysis. Cluster I cells included the full set of undifferentiated progenitors, and Cluster IV cells all the differentiated neurons. Clusters II and III showed highly similar gene expression and were most closely matched with the neuronally-biased ventricular zone progenitors and the basal progenitors, respectively. Clustering revealed that these two groups occupy the same branch of the hierarchical tree as the neurons, setting them apart from the undifferentiated progenitors in Cluster I. Equipped with a clear in vitro picture of neuroprogenitor taxonomy, Kawaguchi next looked in vivo, to see if gene expression patterns would match the predictions of the cluster analysis. In situ hybridization revealed distinct patterns of expression along the apical-basal axis, consistent with the expectation that the Cluster II cells represented progenitors that have begun to differentiate, and Cluster III basal progenitors in the subventricular zone. The high degree of similarity in gene expression between Clusters II and III raised the possibility that the Cluster II cells were young progenitors in the ventricular zone en route to more basal destinations in the subventricular zone, where they would mature and divide to produce a pair of terminally differentiated neurons. Cluster I cells, in contrast, appeared more likely to be undifferentiated apical progenitors, whose nuclei undergo elevator-like shuttling in vivo. Looking at levels of molecules associated with neuroprogenitor fate determination, Kawaguchi et al. found a remarkable heterogeneity in factors in the Notch signaling pathway, although it was unclear whether this was due to transient contact or loss of contact by individual cells with sources of Notch ligands in their microenvironments, or an inherent diversity within the Cluster I population. In any case, Notch signaling provided the strongest distinction between the Cluster I and Cluster II/III groups. Expression of the Notch ligand Delta-like 1 was seen most strongly in the Cluster II cells, suggesting that these young basal progenitors are an important Notch signaling source. Inhibition of this pathway in cultures including Cluster I (apical progenitor) cells resulted in dramatic increases in the number of cells that converted from an apical to a basal progenitor fate. The picture that emerges is one in which young basal progenitors express a ligand capable of inhibiting the differentiation of their neighbors as they make their way to the subventricular zone, thereby maintaining the balance between undifferentiated and committed progenitors. “This is the first work to provide a genome-wide basis for the molecular properties and variety of neural progenitors in mammalian developing brains,” says Matsuzaki. “This allows us to clearly delineate one dynamic feedback system controlling the balance between self-renewal and differentiation among progenitors.” |
||||
|
||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |