| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
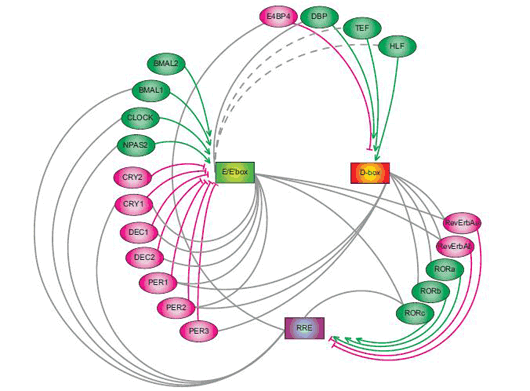
September 30, 2008 – Much like their mechanical counterparts, mammalian biological clocks rely on complex inner workings. In mouse, the circadian clock is established by a system of interlinked genetic regulatory loops consisting of at least three sets of clock-controlled DNA elements that mediate transcriptional output at different times of day: morning, daytime and night. With twenty such elements already identified, the genetic regulation of the biological clock already rivals the most sophisticated clockworks in its complexity, but the dynamic principles that govern this system have never been clearly defined.
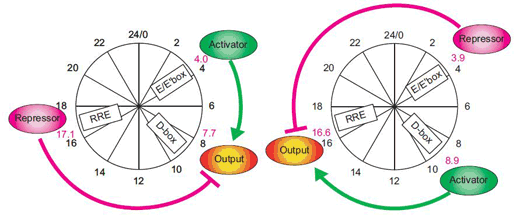
A recent study by Maki Ukai-Tadenuma of the Laboratory for Systems Biology and Takeya Kasukawa of the Functional Genomics Unit (both of which are led by Hiroki R. Ueda; Team Leader/Unit Leader) provides new insights into the transcriptional logic of the mammalian circadian clock. In an article published in Nature Cell Biology, Ukai-Tadenuma and Kasukawa describe how they simulated this circuitry in cellulo using synthetic factors to manipulate and observe the system dynamics of the biological clock. The work began with the development of a clock-cell culture system in which the expression of a synthetic transcriptional activator and repressor were put under the control of morning (E’-box), daytime (D-box) or night (RRE) clock-control transcription factors. By transiently transfecting the host cells with the activator and repressor, along with a reporter construct used to monitor output, the team was able to watch their transcriptional activity in real-time over the course of several days per transfection. Testing a hypothesis that a morning activator combined with a nighttime repressor would be sufficient to produce daytime transcriptional output (which was based on experimental results from previous work in the Ueda lab), they put their synthetic activator and repressor under the control of E’-box and RRE elements, and observations the resulting oscillations in output as indicated by the luminescent reporter. The transcriptional output tracked very closely (within 1 hour) of the rhythm seen in the output of the natural daytime phase controlled by D-box elements. A related principle for nighttime output, determined by a daytime activator and morning repressor, yielded similar confirmation of theory; the transcriptional output of the synthetic circuitry oscillated in near-lockstep with the natural daytime clock, providing an important proof-by-synthesis validation of the logics underlying the regulation of the mammalian biological clock in vivo.
Confident that their synthetic system could faithfully mimic at least two natural daily rhythms, Ukai-Tadenuma and Kasukawa next attempted to generate circadian phases not seen in the natural biological clock. The pairing of a night activator and a daytime repressor, for example, generated high-amplitude transcriptional output in the pre-dawn period, while other combinations of morning, daytime and night activators and repressors likewise resulted in oscillations with regular circadian phases. Given these results, they reasoned that in principle, it should be possible to generate transcriptional output that would peak at times corresponding to (subjective) midday, dawn, dusk or late at night. A previous report from the Ueda lab had proposed an even more general model for circadian oscillations under genetic control. In this model, the expression of a repressor in advance of an activator element (“repressor-precedes-activator”) results in the delay of transcriptional output, while in the opposite case (“activator-precedes-repressor”), the phase is advanced. A corollary to this is that larger discrepancies between the repressor and activator phases (“repressor antiphasic to activator”) create higher amplitude oscillations. Using this as a starting point, they searched for theoretical sets of parameters likely to generate specific outputs, and created an input-output plot against which they could test the theory using their synthetic activator and repressor. As with the morning, day and night rhythms, their experimental data matched nicely with the predictions of the general activator-repressor theory, showing that, in this system at least, the virtual can provide a window into the real, just as manmade clocks keep us informed of the natural passage of time. “Physical simulations allow us to mirror and manipulate actual biological systems with increasing precision and ease, making it possible to perform all kind of analysis,” says Ueda. “This approach should be equally valid in the study of dynamic, complex biological systems other than biological clocks as well, so we are hopeful that it will find a broad range of applications.”
|
|||||||||
|
|||||||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |