| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
April 18, 2010 – Maintaining the integrity of the genome is a mission-critical role for the cell’s many DNA repair mechanisms. Errors of varying severity can creep into the code in the form of substitutions, deletions, reversals and extra copies, but ruptures of the double helix itself can be even more catastrophic. Such double-stand breaks set off alarms, as they can lead to massive rearrangements of long stretches of the genome, and eukaryotes have evolved an arsenal of detection, repair, and abort mechanisms to deal with this contingency. Homologous recombination is one tactic used to salvage broken DNA, by making a template for the damaged sequence from a sister chromatid; a very similar process takes lace during meiosis.
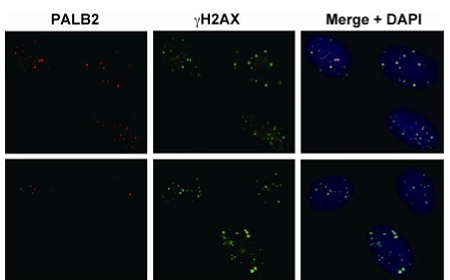
Studies of homologous recombination have revealed a number of the molecular factors that underlie the process, with the genes BRCA1 and BRCA2 (whose loss of function is a well-known predictor of breast cancer) playing central roles. The proteins encoded by these genes are bound together in a complex with other factors known as RAD51 and PALB2, which respectively form single-strand filaments of DNA and protein that are used in initiating the recombination event, and help recruit BRCA2 to chromatin containing damaged DNA. The chromatin-side target of the BRCA complex, however, remains unclear. A new study by Tomohiro Hayakawa of the Laboratory for Chromatin Dynamics (Junichi Nakayama; Team Leader), Fun Zhang in Paul Andreassen’s lab at Cincinnati Children’s Research Foundation and others has added a new link to that chain, showing that the chromodomain protein MRG15 binds directly to PALB2, and is required for homologous recombination. Published in The Journal of Cell Science, this new work provides new insight into how the DNA damage response network interacts with the genome through its nucleoprotein packaging. Knowing that a molecular relative of MRG15 complexes with PALB2, Hayakawa and Zhang started by testing for a possible functional link using immunoprecipitation, and found that the two proteins show affinity for each other. Further experiments using Western blotting revealed that MRG15 indeed interacts with the entire BRCA complex. Testing next for function, the team found that MRG15 plays a vital role in homologous recombination; depletion of the protein caused a significant drop in the efficiency of this repair mechanism. Interestingly, MRG15 also appeared to be tied to the activity of mitomycin C (MMC), a DNA crosslinker protein that can affect replication. MMC resistance is mediated by the BRCA complex, and Hayakawa and Zhang found that loss of MRG15 function caused hypersensitivity to the crosslinking agent, similar to the phenotype when either BRCA2 or PALB2 are knocked down. The recruitment of the BRCA complex to the site of DNA damage has been shown to rely on PALB2, so the researchers looked for a possible role for MRG15 in this targeting. Using immunoblotting to look at BRCA complex binding to chromatin in wild type and irradiated cells in which MRG15 was expressed normally or silenced by siRNA, they found that the DNA repair complex was associated with chromatin at much higher frequencies in untreated cells (see Fig.). They speculate that once PALB2 has attached to MRG15 in the chromatin, it then recruits the other elements of the BRCA complex, enabling homologous recombination to proceed. “Several lines of study have shown how dramatically chromatin structure can change over the course of development,” says Hayakawa, “but we still know very little about what roles MRG15, PALB2 and the BRCA complex might play in such rearrangements. We’re hopeful that by studying these interactions we will gain a better understanding of developmental processes.”
|
||||
|
||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |