| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
December 23, 2010 – Cells have evolved a variety of strategies for ensuring that they get just the right doses of gene products. X inactivation, in which one of the two X chromosomes in female mammals is epigenetically silenced, is one of the best known and most intensively studied of these mechanisms. One of this process’s more interesting features is that cell lineage determines whether inactivation occurs in a randomized or deterministic fashion. In somatic lineages differentiated in post-implantation embryo, X inactivation can take place in either the paternally or maternally supplied chromosome, while in extraembryonic tissues segregated from pluripotent stem cells in pre-implantation embryo, the paternal X is always inactivated. But has this transition already been made in the pluripotent stem cells of inner cell mass, which represents both the first fruit of differentiation and the starting point for all somatic lineages?
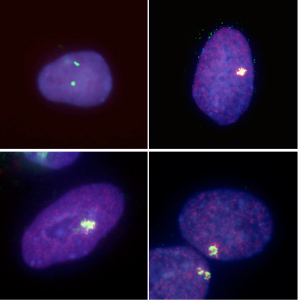
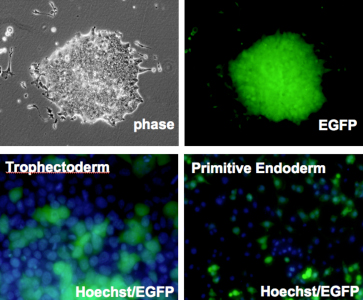
Kazuhiro Murakami in the Laboratory for Pluripotent Stem Cell Studies (Hitoshi Niwa, Project Leader) and colleagues used a pair of modified mouse embryonic stem (ES) cell lines to answer that question, and found that when the pluripotent cells were forced to differentiate into extraembryonic lineages, X inactivation in their progeny cells took place randomly. Their findings, published in Development, help to better constrain the timing of this crucial epigenetic event. The team selected two lines of female mouse ES cell lines in which the paternal and maternal X chromosomes could be readily distinguished, due to genomic polymorphism or fluorescent labeling. ES cells such as these do not normally give rise to extraembryonic lineages, such as trophectoderm or primitive endoderm, but previous work by the Niwa lab had shown that forced expression of Cdx2 or Gata6 could steer ES cells down those respective pathways. Murakami used this combination of cells and transcription factors to generate ES-derived extraembryonic lineages in which the pattern of X inactivation could be visualized. X inactivation is associated with the epigenetic silencing by trimethylation of histone 3 lysine 27 (H3K27) as well as the coating with Xist RNA, so the team first confirmed that their induced trophectoderm and primitive endoderm cells had indeed undergone silencing, and found that, in contrast to undifferentiated ES cells in which inactivation has not occurred, the extraembryonic cells showed clear Xist-H3K27 association at a single locus. The question was: did this happen only in the maternal X, or randomly to either the paternally or maternally donated chromosome? Murakami et al. first used a PCR-restriction fragment length polymorphism (RFLP) assay to detect transcripts specific to each genomic donor. While both sets of transcripts are expected to appear in undifferentiated cells, only the maternal transcripts appear in normal extraembryonic tissues, due to imprinted X inactivation. However, in the induced trophectoderm and primitive endoderm (in which X inactivation clearly occurs), both paternal and maternal X chromosome contributions remained, suggesting that the process occurred randomly.
This finding was supported by results of parallel experiments using a second ES cell line in which the paternal X chromosome was labeled by EGFP. If imprinted X inactivation took place in the induced extraembryonic cells, the fluorescent signal would be lost, but in both primitive endoderm and trophectoderm, the cells glowed, testifying to the stochastic nature of the chromosomal silencing. To make a final confirmation that the X inactivation imprint is already lost in the inner cell mass, the team, in collaboration with the Laboratory for Genomic Reprogramming (Teruhiko Wakayama, Team Leader) used the ES cells expressing EGFP on its paternal X chromosome to generate cloned blastocysts. As anticipated by the results from their in vitro differentiation studies, X inactivation in the trophectoderm of these blastocysts occurred randomly, in contrast to the usual imprinting seen in the same tissue in control embryos. The imprinting that leads to selective X inactivation, it seems, is completely lost in ES cells. “The precise developmental time-point of the transition in the X chromsome’s epigenetic state from imprinted to random X inactivation was unknown, but we were able to show that the early epiblast from which ES cells are derived completes the transition event by erasing all epigenetic modifications for imprinted X inactivation,” says Niwa. “The epigenetic state of ES cells may be a sort of tabula rasa.” |
|||||||
|
|||||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |