| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
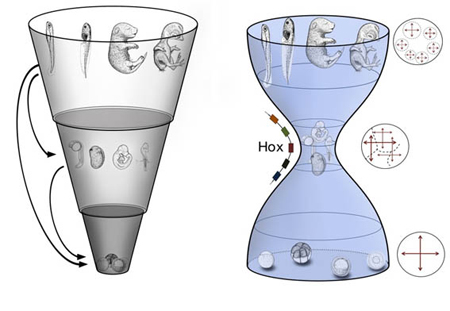
April 10, 2011 –The concept of the phylotypic stage traces its roots back to early comparative observations of embryos from different vertebrate taxa, in which it was noted that embryonic morphologies appeared to converge on a shared body plan before veering off in specialized directions. This gave rise to a profound debate over the evolutionary basis for this phenomenon; specifically, whether it could best be explained by a “funnel” model, in which the commonality of traits is highest at the earliest stages of embryogenesis, and gradually but unilaterally narrows over time, or an “hourglass” model, where homology is highest at a point later in development as the body plan is being established, and differs more widely before and after.
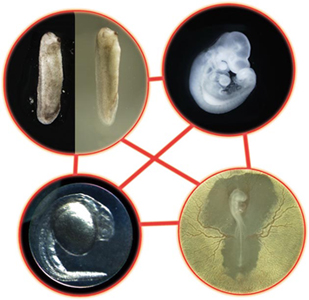
A new comparative transcriptomic analysis of four vertebrate species conducted by Naoki Irie in the Laboratory for Evolutionary Morphology (Shigeru Kuratani, Group Director) has now revealed that genetic expression is most highly conserved across taxa at the pharyngula stage of development. Published in Nature Communications, these latest findings strongly suggest that the hourglass model is the more accurate description of how the vertebrate phylotype manifests. Irie decision to study this question using a gene expression approach broke with the long history of morphological comparisons. He sampled tissue from mouse, chicken, and frog embryos across multiple developmental stages to allow for comparisons of changes in gene expression, and further supplemented this data set with information from previously published transcriptomic studies in a fourth taxa, zebrafish, thus providing representative samples from mammal, bird, amphibian and fish species. He took advantage of the supercomputing capabilities at the RIKEN integrated Cluster of Clusters (RICC) for the processing power needed for comparison of this enormous set of data points. As development proceeds at different paces in different species, and organs likewise emerge at different points, making straightforward comparisons can be extremely problematic. To overcome this challenge, Irie selected genes homologous in all four species, and made pairwise comparisons of gene expression profiles by microarray analyses. For each pairing, he found that the highest similarity was seen in intermediate stages of embryogenesis (from neurula to late pharyngula stages). More sophisticated computational analysis revealed that pharyngular embryos had the highest transcriptomic similarity of any stage. To ascertain the detailed molecular characteristics of this phylotypic stage, the group went on to identify genes showing conserved expression during the pharyngula stage, but that were not constitutively expressed throughout embryogenesis, and identified 109 gene sets, including Hox genes, transcription factors, cell-cell signaling genes, and morphogens. Interestingly, within these sets, developmental genes were more highly represented than in sets with different expression profiles. The data generated in this study has been deposited in the ArrayExpress and Gene Expression Omnibus repositories.
“It seems that the notion that genetic programs underlying early development are resistant to change needs to be reconsidered in light of this data,” says Irie. “We’ll be interested in working out how early genetic flexibility is achieved while maintaining the robustness of gene expression at the phylotypic pharyngula stage.” | |||||||
|
|||||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |