| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
April 15, 2011 –The separation of the germ cells responsible for the transmission of genetic information across generations through reproduction from the somatic tissues that form the rest of the body is a critical event in early development. In many organisms, germ cells are characterized by the inclusion of organelles known generally as germ granules, whose function appears to be linked to germ cell specification and differentiation. In the nematode C. elegans, these organelles are historically known as P granules, and are made up of a combination of messenger RNAs and protein components, only some of which have been identified.
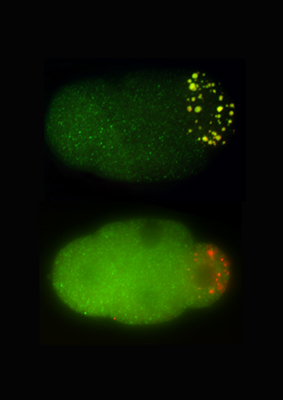
Momoyo Hanazawa and Masafumi Yonetani in the Laboratory for Developmental Genomics (Team Leader, Asako Sugimoto; now at Tohoku University) have now sorted out a crucial role for a pair of proteins in germ granule formation. The study, published in the Journal of Cell Biology, shows how PGL-1 and -3 self-assemble into scaffolds that recruit ribonucleoprotein components to developing granules. The team began by developing an assay system to test individual components of P granules for the ability to form granules independent of other granule-associated proteins in a mammalian cell-based system. Of the fourteen proteins tested, only two – PGL-1 and PGL-3 – assembled into granular clusters. To test this capacity in C. elegans cells, Hanazawa induced the expression of these PGL proteins in embryonic and adult somatic cells and confirmed the formation of granules in these cellular environments as well, suggesting that these factors are able to self-organize into structure that may help to recruit other P granule components. To test this notion, they next checked whether RNAs and proteins co-localized with the PGL complexes. They found that the granules stained positive by a ribonucleic acid-specific dye, and that eight of 12 granule proteins tested also co-expressed with PGL-3. They next used a loss-of-function approach in which they depleted the PGLs from C. elegans embryos using mutations or RNAi, and determined that granule formation was greatly reduced in the PGL-deficient animals. This was true for members of the GLH family (homologs of VASA in fluitfly), which typically co-localize with PGL granules in germ cells as well. Analysis of cells in which both GLH-1 and -4 were knocked down below the detection threshold showed that while PGL proteins were still able to self-aggregate, their GLH partners were required for maintaining the multicomponent granular structures. Yonetani and Hanazawa next performed a structural analysis of PGL-3 to identify possible functional domains. They found two domains are essential to form germ granules: one is an RNA binding domain in the protein’s C-terminal, which appeared to be essential for the capture and incorporation of RNAs to granules, and the other a self-association domain in the N-terminal, which is essential for forming globular granules. These findings suggest that the self-association of RNA binding proteins, such as PGL proteins, is crucial for forming germ granules containing diverse RNA and proteins. Sugimoto notes, “There are many RNA-and-protein containing granules (ribonucleoprotein granules, or RNP granules) in a cell, which play important roles in gene expressions. Our finding may provide a general mechanism for the formation of RNP granules.” | |||||
|
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |