| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
February 24, 2012 ĖThe mammalian brain is one of the pinnacles of evolutionary achievement; myriad interconnecting neurons in discrete functional domains give rise to an organ of astonishing complexity. Numerous efforts have been made to profile the molecular signatures of the brainís many regions, typically using a technology known as in situ hybridization to visualize gene transcripts at specific sites. Other more comprehensive techniques for analyzing gene expression, such as DNA microarrays, also exist, but these have yet to be used in measuring the central nervous systemís genetic activity across a wide range of different regions. Takeya Kasukawa and Itoshi Nikaido of the Functional Genomics Unit, Koh-hei Masumoto of the Laboratory for Systems Biology (both headed by Hiroki R. Ueda) and others have done just that, profiling genome-wide expression across nearly 50 brain regions in the mouse. Published in PLoS One, this work identified a large number of genes showing interesting characteristics such as multi-state expression, control properties, and putative ligand-receptor interactions. The results have been compiled into an open, searchable database at http://brainstars.org.
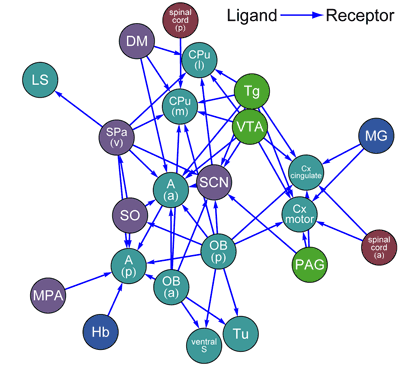
The group sampled 51 brain regions associated with distinct functions, such as various sensory modalities, internal clocks, memory, fear, and feeding, at 4-hour intervals over the course of a day. They next used quantitative PCR (qPCR) to analyze these samples for the expression of region-specific genes, and used global clustering to identify developmental, evolutionary and anatomical groupings, noting a high degree of consistency with expectations, but a few notable exceptions as well. Retina, pituitary and pineal tissue, for example, were markedly distinct from the other 48 regions in this analysis, which may be due to differences in anatomy or cell-type composition. Their next step was to identify and analyze multi-state patterns of gene expression, finding more than 8,000 genes showing multimodal expression across the sampled regions. Such multiple states may reflect either differential regulation in different cell types, or heterogeneity in the balance of various cell types in different brain regions. A closer examination revealed a number of interesting genes with from two to four distinct states, including various G-protein-coupled receptors (GPCRs) and a large proportion of homeobox and nuclear receptor genes. Such multi-state expression data was then used to identify candidate marker genes, defined as those whose highest or lowest expression state occurs only in a single region of the central nervous system. More than 2,000 such genes were found, including several transcriptional regulator/target gene pairs. Not all genes are multimodal, however; the study also found a class of genes with single-state expression across multiple CNS regions, suggesting that they may function as stable internal controls. To test the utility of their dataset, Kasukawa and Masumoto focused on neurotransmitter (NT) and neurohormone (NH) ligands and receptors as a means of measuring the interconnectivity between CNS regions, by counting the number of NH/NT ligand-receptor expressions for every pairing of CNS regions, and plotted these by density. In examples of the revelatory power of this approach, they identified a high level of connectivity between the hypothalamic and olfactory bulb regions, and large number of inbound connections to the suprachiasmatic nucleus, the circadian clock center, suggesting the requirement for multiple inputs in keeping the bodyís internal clock running on time. A number of other intrinsic connections, described in detail in the article and the public database, were also uncovered. The final validation of the groupís findings came from a comparison to several previous studies of regional gene expression in the mouse brain. Despite the smaller sampling sizes of the BrainStars study, it showed good levels of correlation with existing in situ hybridization data sets, with some differences however in findings for smaller and more complex regions, such as the hypothalamus. "The BrainStars database provides a range of information on gene expression in the mouse brain, including gene expression profiles the results of various analyses," says Kasukawa. "We are hopeful that this resource will make a contribution to CNS research, and would be delighted to see it used widely by others." |
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |