| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
April 10, 2012 –The enormous information set encoded in a cell’s genome is bundled into tightly wound coils of DNA and proteins known as chromatin. The prevailing view has been that this structure takes two main forms: loosely packed stretches of euchromatin which tend to permit active transcription, and more securely locked down heterochromatin, where gene expression is typically silenced, and which provides the structural basis for regions such as the centromere and telomeres. Heterochromatin is the end product a cascade of events involving multiple molecular complexes that interact to form this critical chromosomal component, which is characterized in part by its coating of the protein HP1. Some studies have suggested that HP1 plays a role upstream in the cascade as well, but the details of this have resisted explication.
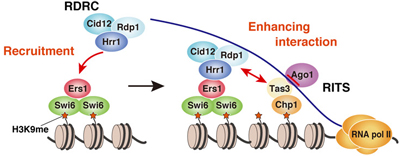
Now, Aki Hayashi and others in the Laboratory for Chromatin Dynamics (Jun-ichi Nakayama, Team Leader) have shown that the co-factors Ers1 and HP1 homolog Swi6 work together to recruit the RNAi machinery to sites of heterochromatin assembly in fission yeast, Schizosaccharomyces pombe. Published in the Proceedings of the National Academy of Sciences, this work refines our understanding of how this crucial basis of chromosome structure is formed. Heterochromatin assembly is regulated by Histone H3 Lysine 9 (H3K9me) methylation. In fission yeast, the histone methyltransferase Clr4 marks specific sites on chromosomes, serving as a binding site for chromodomain proteins such as Swi6, which in turn induces heterochromatin compaction. Interestingly, the molecular machinery behind RNAi, best known as an epigenetic form of post-transcriptional regulation, also plays a part. In S. pombe, the RNA-induced transcriptional silencing (RITS) complex and RNA-dependent RNA polymerase complex (RDRC) both function in the RNAi process, but recent work has shown that these are, surprisingly, also essential to heterochromatin assembly in the centromeres, as loss of function of either results in aberrant centromeres and defective gene silencing. This growing body of work has shown that both RITS and RDRC localize at centromere heterochromatic regions, highlighting the possibility that this RNAi machinery is involved in heterochromatin assembly. Hayashi approached this question using a genetic screen for mutations affecting gene silencing at the centromeres, which yielded a number of hits associated with the RNAi machinery, including a factor called Ers1, which had previously been necessary to RNAi, but remained poorly understood. The team found that ers1 mutants showed not only defective silencing, but also low production of siRNAs (short interfering RNAs) and reduced H3K9 methylation. On testing the ability of various genes involved in heterochromatin assembly or RNAi to rescue these phenotypes, they found that overexpression of RNA helicase Hrr1 and the methyltransferase Clr4 had a partially compensatory effect, pointing to the possibility of a functional connection. Yeast two hybrid assays showed that Ers1 and Hrr1 bind directly, but did not show any physical association between either Ers1 or Hrr1 with Clr4. Given these findings, questions remained regarding the molecular mechanisms behind the functional association. Hayashi et al. next used EGFP to label Ers1 in wildtype cells, allowing them to observe its punctate localization in the nucleus, consistent with co-localization with heterochromatin. In Clr4 and Swi6 mutants, however, the localization was more diffuse, indicating their importance to the localization of Ers1 to heterochromatic regions. A subsequent yeast two-hybrid screen confirmed that Ers1 binds directly with Swi6, fitting a piece into a puzzle that had begun clarifying into a scenario in which the molecular complex of Ers1 and Swi6 is recruited to heterochromatin regions methylated by Clr4. The bigger picture reveals that Ers1 interacts with both Hrr1 and Swi6. Hrr1 is a necessary component of RDRC, which the team found was recruited to centromeres by Ers1 and Swi6. Thus, Swi6 acts as a kind of platform to localize this complex to these highly methylated heterochromatic regions in order to proliferate further siRNA generation. “Recent work has revealed the direct association between the RITS and RDRC complexes,” says Nakayama. “It seems that Ers1 is playing a critical role in bridging the RNAi machinery with heterochromatin assembly. It is will be very interesting to see whether this bridging mechanism, as well as the mechanism behind RNAi mediated heterochromatin formation, are conserved in other organisms. ” |
|||||
|
|||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |