| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
Feb 25, 2013 – Confocal microscopy is a common tool for looking at the interiors of living samples in three dimensions. To increase the speed of image acquisition, multi-point scanning methods using a spinning disc embedded with thousands of pinholes that remove out-of-focus background light have been developed. But while spinning disk confocal microscopes cause less radiation damage to samples and achieve higher temporal resolution than conventional alternatives, the conventional model was limited in its ability to probe the interiors of large specimens due to a phenomenon known as ‘pinhole crosstalk,’ in which out-of-plane and scattered emissions bleed through inappropriate neighboring pinholes and increase the background haze that obscures images. A new development by Togo Shimozawa and others in the Optical Image Analysis Unit (Yuko Mimori-Kiyosue, Unit Leader) shows a way out of the crosstalk by adjustments to pinhole placement and the use of two-photon excitation to reduce out-of-focus light. Published in the Proceedings of the National Academy of Sciences, these enhancements extend the range of applications of spinning disk confocal microscopy in the live imaging of larger specimens, including whole embryos. Shimozawa has since moved to Gakushuin University.
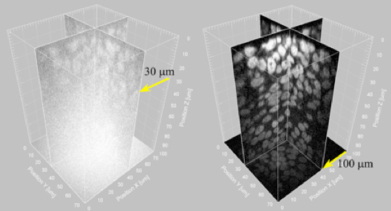
Expecting, based on theory, that the problematic pinhole crosstalk might be eliminated by increasing the space between these apertures, the lab modified a standard Confocal Scanner Unit (CSU), doubling the distance between pinholes as well as the size of the microlenses that focus light onto them. The doubling of microlens size increases the concentration of light to the pinholes, boosting two-photon absorption efficiency, which is crucial to eliminate background light generated in out-of-focus planes, a key contributor to pinhole crosstalk. A comparison of the two-photon excitation (2PE) setup with the standard CSU revealed the benefits of the new approach. While in the conventional technology background noise due to crosstalk can be as high as 60% of the total detected signals when imaging a thick specimen, with 2PE this dropped to as low as 0.5%. When they used the new apparatus to image a bulky (diameter 30 µm) pollen grain, not only was background light reduced, but 2PE also revealed rich new features of the particle’s interior. And by assembling multiple images taken in this fashion, the lab was able to reconstruct its structure as a 3D image, puling out details from even its far side that remained obscured in the background of single-photon images. For their next demonstration, Shimozawa et al. used the 2PE system to examine GFP-expressing model organisms of the sort typically used in embryology studies. In mouse embryos, where traditional spinning disk confocal microscopes can only visualize to depths of around 30 µm, the new system could image three times as deep, to over 100 µm. The 2PE unit also showed the ability to visualize submicron structures in vivo. Looking at Drosophila embryos, they were able to follow the movements of a microtubule plus end-associated protein, EB1, by time-lapse imaging. “We’re very hopeful that this new technology will find applications across the life sciences and in other disciplines as well,” says Mimori-Kiyosue. “But as the power of the lasers that generate the multiple beams needed for this approach is the limiting factor, we are looking to develop higher-power laser sources that will allow us to look even deeper and more clearly into microscopic specimens.” |
|||||||
|
|||||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |