| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
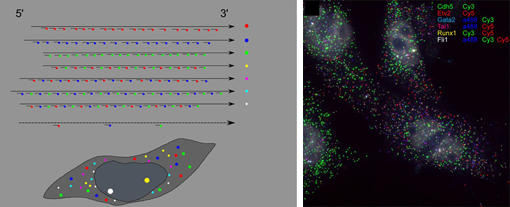
Mar 28, 2013 –Techniques for imaging the inner workings of cells are a backbone of modern biological investigations. In developmental biology studies, information on the behavior and properties, such as gene expression, of interacting cells is fundamentally important for gaining insight into their respective spatial arrangements and dynamics. Fluorescence in situ hybridization (FISH) is a well-known method for visualizing the localization of individual RNA molecules within cells, providing insight into the expression of genes of interest, but this technique is limited in the number of different genes it can be applied to simultaneously in a single cell. In a new report, Lars Martin Jakt and colleagues in the Lab for Stem Cell Biology (Shin-Ichi Nishikawa, Group Director) describes a new variant of this technique that uses large numbers of short probes making it possible to target transcripts by multiple fluorescent molecules, dramatically expanding the number of genes that can be visualized in situ. Published in Development, this new CandyFISH method may open the way to the simultaneous visualization of transcripts from 10 or more genes. In a proof of concept demonstration Jakt, now at Keio University, put the technique to use, revealing changes in gene expression in individual cells during the differentiation of mesoderm into endothelium.
The original FISH method involves labeling mRNA molecules with fluorescent RNA probes, enabling the visualization of the types and amounts of expressed genes in living cells. A recent improvement to this method have fused multiple short probes to single genes, yielding higher fluorescence and comparatively low cost, but with the limitation of being applicable to a maximum of only three genes. Jakt’s refinement shows that using many short probes allows transcripts to be targeted by multiple fluorophores, allowing the quantification of gene expression from more than double that number of genes. In a proof of principle experiment, the group next used the new method to examine changes in gene expression profile of cells during the differentiation of vascular endothelium from embryonic mesoderm. Previous in vitro studies had shown Etv2 to be an important gene in this process, involved in the induction of endothelial genes, as well as a direct inducer of genes involved in stabilizing the hematopoietic state. This diversity of downstream targets made for a very fuzzy picture of the order in which Etv2 activates other genes. Jakt studied ES cells induced to differentiate into endothelium using six genes of interest marked using the CandyFISH technique, enabling him to visualize the intracellular localization of their RNA transcripts. By plotting changes in transcript density in individual cells over time, the group was able to work out a progression of the switching on and off of Etx2 and its targets, yielding a much clearer image of how gene expression changes in single cells during endothelial differentiation. CandyFISH represents an important new step in simultaneously measuring the abundances of multiple RNAs at the single-cell level, and consequently tracking changes in cellular identity. Additional refinements in microscopy and fluorescent labels may enable the tracking of the expression of as many as 10 genes in individual cells, and to analyze deeper tissue strata. |
|||||||
|
|||||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |