| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
Apr 26, 2013 – Sleep is a crucial daily activity for an extraordinarily wide range of animal species. Consciousness is typically divided into waking, REM (rapid eye movement), non-REM states, reflecting patterns of activity recorded in electroencephalography (EEG) and electromyography (EMG) data. Analyses of sets of such data are indispensable for studies of the mechanisms underlying sleep/wake states and sleep pathologies, but the need for inspection by individual human raters has imposed limits on the scale and speed of assessment and introduced problems of standardization and potential subjective biases. A new automated approach to sleep staging developed by Genshiro A. Sunagawa and colleagues in the Lab for Systems Biology (Hiroki R. Ueda, Project Leader) now potentially makes the process more scalable and objective. Published in Genes to Cells, this new FASTER (Fully Automated Sleep sTaging method via EEG/EMG Recordings) method was developed in collaboration with researchers from Tokushima University, Osaka Bioscience Institute, and Nihon University College of Pharmacy.
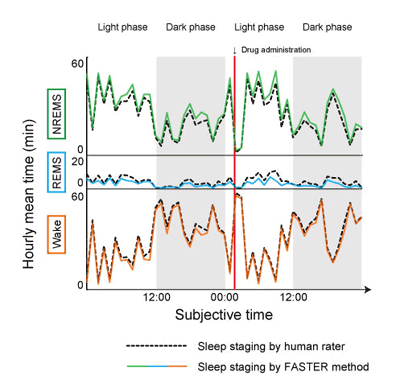
The EEG and EMG waveforms observed during sleep differ between individuals, and between strains of laboratory mice. For this reason, sleep analyses have traditionally relied on the rating of data obtained from individual animals over a fixed period (such as 24 hours) to establish standards for the three sleep/wake stages. This process requires individual raters to take into consideration inter-individual differences and eliminate noise, but also introduces subjectivity and the risk of human error, as well as limiting the scale of data that can be evaluated. Semi-automated approaches have been developed, but these still require human supervision over the setting of scoring criteria. The FASTER method developed by Sunagawa and colleagues now provides a fully automated alternative. The algorithm is composed of three main steps: character extraction, clustering, and annotation. The first of these involves breaking the observation period into epochs of fixed length (8 sec), converting the EEG/EMG outputs for each epoch into frequency domain data, and then subjecting them to principal component analysis to extract characters of their power spectra. After clustering regions showing similar features as determined by nonparametric density estimation, the annotation was performed without human supervision as determined by statistical characteristics for each cluster. The team next tested the FASTER algorithm against results from human rater evaluations of mouse sleep/wake stages in wildtype animals in which these patterns were altered by drugs that induced wakefulness or sleep. When they compared the results of human raters’ assessments with those generated by the FASTER algorithm, they found greater than 90% agreement between the two, indicating the high level of precision of this unsupervised, fully automated method. The FASTER approach was also...faster, taking only 10 minutes to complete an analysis that would require 1–2 hours using conventional methods.
“FASTER by itself won’t make it possible to automate the staging of huge sleep data sets,” cautions Sunagawa. “There is still quite a lot of manual work involved, such as attaching electrodes to mice, something we hope to tackle in future work. We are also looking forward to applying this new approach to sleep staging in humans, which will require handling much larger amounts of data.” |
|||||||
|
|||||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |