| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
Dec 5, 2013 – Chemotaxis refers to movement by cells to or away from chemical stimuli in their environments. In the multicellular body, this process plays a central role in directing immune responses by white blood cells and the formation of nervous system tissues. For more primitive eukaryotes, such as the slime mold Dictyostelium, chemotaxis plays what is perhaps an even more fascinating role, guiding individual cells in food-poor environments to assemble into a multicellular aggregate, or ‘slug,’ capable of reproduction. The mechanism underlying this chemotaxis resembles that seen in mammalian immune cells, and allows tiny cells just a few micrometers in diameter to respond to minute differences of a few percent in the concentration of a chemical stimulus. However, cells do not need an excuse to move; even in the absence of a chemotactic signal, cells move in random direction under the control of internal fluctuations. The question then is, how do cells override these random internal signals so as to be able to detect, amplify, and respond to gradients of external stimuli? In a report published in Journal of Cell Science, Team Leader Tatsuo Shibata and colleagues in the Lab for Physical Biology used fluorescence imaging to reveal a mechanism for controlling the spontaneous polarization of Dictyostelium cells, from which they developed a quantitative model able to reconstruct the phenomenon. A second paper by the same lab, published in Biophysical Journal, reports a model of how this self-organizing mechanism robustly facilitates the exquisite sensitive of these cells to chemical stimuli. Both studies were conducted in collaboration with researchers from the RIKEN Quantitative Biology Center.
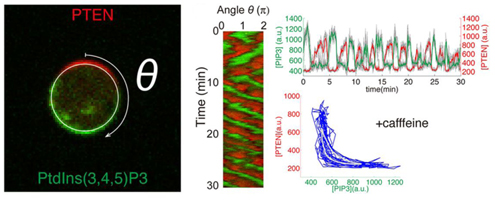
Dictyostelium cells respond chemotactically to gradients of cAMP by initiating inositol phospholipid metabolism at the cell surface. Cell surface regions presented with a high cAMP concentration begin to produce the phosphatidylinositol lipid PIP3 from PIP2, while regions facing lower concentrations elicit the opposite reaction, catalyzed by the enzyme PTEN. Regions of high PIP3 mobilize actin fiber formation to extend a pseudopod in the direction of the gradient, driving chemotaxis. Shibata wondered, however, how would cells behave in environments lacking cAMP, and simultaneously prevented from generating cAMP themselves. He found that PIP3 domains appeared to move at random on the surface of such cells, indicating an internal mechanism for establishing polarity, ensuring that PIP3 and PTEN localized in a mutually exclusive fashion. Statistical analysis revealed that this activity could not be explained simply as the result of interactions between enzymes and substrates, but that rather PTEN seemed to be ‘attracted’ to areas in which PIP3 levels were low. Using a reaction–diffusion model to simulate this phenomenon, Shibata et al. were able to show how domains of persistent PIP3 expression, in agreement with their experimental observations. Adjusting the simulated parameters, he was able to evoke cycles of PIP3 oscillation, resembling the behavior of cells in which cAMP was not blocked. These validations of this quantitative model point to possible broader applications in the analysis of spontaneous polarization, such as seen in neurons, which can exhibit robust responses to even seemingly minor stimuli.
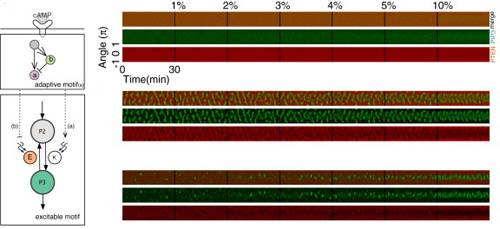
In the second study, the team looked at cells confronted with chemotactic stimuli. Using ordinary Dictyostelium cells, they observed that while some would manifest stable domains in response to cAMP, others showed transient domain patterns oriented toward higher cAMP concentrations. They next made a stochastic reaction–diffusion model of these two patterns, along with that of an inactive state absent the self-organized polarity formation, and tested how they would respond to stepwise increases in cAMP gradient steepness. The persistent and transient domain models responded much more strongly than did that of the inactive state, indicating that the self-organizing system plays a role in promoting the response to cAMP. They next turned to quantitatively analyzing PIP3 domain formation, and found that such domains formed more frequently in the presence of a stimulus and became distributed with increasing precision in the direction of the gradient as the chemotactic signal rose. In following cAMP gradients, Dictyostelium cells need to move toward increases in concentration, and avoid pursuing lower levels of cAMP. Shibata suspected that this might be possible through the combined effect of the extracellular gradient, which introduces a small bias in the PIP3 domain induction on the side of the cell facing the higher concentration, and self-organizing activity which establishes front–back orientation. In line with his predictions, the model showed that the self-organizing system lies behind the robust, sensitive response by these cells to even shallow gradients. “In these studies, we were able to construct quantitative models that closely recapitulate the results of experiments using known molecules observed by fluorescence imaging,” says Shibata. “These models have led to new insights into molecular mechanisms and gradient sensing, which we next hope to validate using live cells. We are also hopeful that the self-organizing activity we have described will play a role in the explication of other biological functions.” |
|||||||
|
|||||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |