| RIKEN Center for Developmental Biology (CDB) 2-2-3 Minatojima minamimachi, Chuo-ku, Kobe 650-0047, Japan |
February 10, 2012 –Peptides of the insulin family are found across a broad range of taxa spanning both vertebrates and invertebrates, in which they play roles in the regulation of processes such as metabolism, growth, reproduction and longevity. The genome of the fruit fly Drosophila melanogaster includes seven genes encoding such peptides, known as dilps (Drosophila insulin-like peptides), which are primarily expressed in secretory insulin-producing cells (IPCs) within the brain. The expression of each dilp is independently regulated, but just how this level of coordination is achieved remains poorly understood.
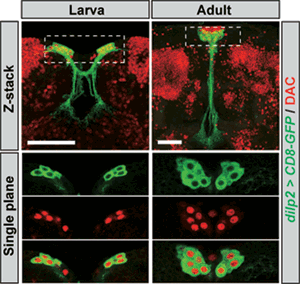
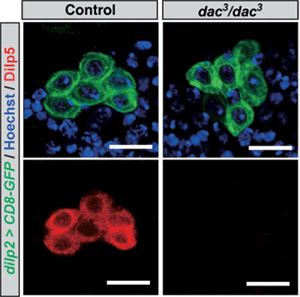
Now, Naoki Okamoto and others in the Laboratory for Growth Control Signaling (Takashi Nishimura, Team Leader) have identified an important clue to this puzzle, showing that a highly conserved nuclear protein Dachshund (Dac) acts as a transcriptional regulator specific to dilp5. Published in the Proceedings of the National Academy of Science, these findings may open new avenues of understanding into the means by which diverse insulin-family peptides are kept working in harmony. The IPCs of the fruit fly brain express several genes known to be involved in the development of the eye. The team knocked down the function of several of these genes in a tissue-specific manner to check for possible effects on dilp expression. They found that the knockdown of dac was accompanied by a dramatic down-regulation of dilp5, suggesting that Dac might function in Dilp5 regulation in IPCs. An examination of Dac expression in IPCs showed the protein is expressed continuously throughout development. When Okamoto next analyzed homozygous mutants for dac, he found that while IPCs formed normally, the expression of dilp5 in these cells was markedly lower in young larvae, but intriguingly its expression was normal in later larval stages, suggesting stage-dependent differential mechanisms for its regulation. A subsequent analysis using heterozygous dac mutants revealed that expression levels of that gene correlated closely with the expression of dilp5. A second protein called Eyeless (Ey) is known to control dilp5 expression, and he found that ey mutants revealed similar phenotypes as found in the dac studies. The team next focused on possible interactions between Dac and Ey, but found that the loss of function of one had no discernible effect on the other, suggesting their expressions are independently regulated. Their effects on dilp5 were synergistic, however, as shown by tests in which the expression of both was perturbed. Looking next for an interaction between Dac, Ey and the dilp5 promoter, they found that while Dac alone did not interact, the binding of Ey to the dilp5 promoter was accelerated in the presence of Dac. Further experiments revealed Dac forms a physical complex with Ey and with itself through specific protein domains. Knowing that Dac is evolutionarily conserved in mammals, Okamoto next turned to the insulin-secreting β-cells in the islets of Langerhans of the pancreas, in which Dach and Pax6, homologs of the Drosophila Dac and Ey, play important developmental roles. Importantly, Pax6 is also involved in the transcription of genes encoding critical pancreatic hormones, such as insulin and glucagon. Using cultured rat β-cells-derived cell line, they examined the functions of these two regulatory proteins and found that, as in the fruit fly, Pax6 and Dach had similar combinatorial effects on the activation of insulin expression.
“The expression of dilp5 can change in response to nutritional status, and we still don’t know how this gene is regulated in later stage larvae,” says Nishimura, “suggesting that the regulatory situation is even more complex than we expected. We are looking forward to tackling the link between nutrition and dilp5 expression in future work.” |
|||||||
|
|||||||
 |
| Copyright (C) CENTER FOR DEVELOPMENTAL BIOLOGY All rights reserved. |