
News and Announcements from the CDB
Targeted genome-editing technology has generated great interest for biomedical research fields for its potential in clinical applications to treat or improve symptoms of some genetic diseases. In particular, the development of the CRISPR/Cas9 system has revolutionized the biology field with the ease and precision in which the genome can be manipulated. This innovative system exploits the activity of CRISPR/Cas9, the bacterial immune system used to respond to pathogen invasion, and the cell’s intrinsic DNA repair mechanism, either via non-homologous end joining (NHEJ), which is seen in both proliferating and non-proliferating cells, or homology-directed repair (HDR), which is used only in proliferating (dividing) cells. Using CRISPR/Cas9 in combination with NHEJ-mediated or with HDR-mediated methods have proven useful for knocking out function of genes through errors caused by small DNA sequence insertions or deletions at target sites during NHEJ repair or specific sequence modifications in gene of interest. In contrast, gene knock-ins have been more challenging, as this usually utilizes the HDR-mediated mechanism, which is unavailable in differentiated cells. Thus, if the NHEJ mechanism, which is found in both proliferating and non-proliferating cells, can be exploited for efficient targeted gene integration, it would pave the way for potential use in vivo, where the majority of cells are non-proliferative.
Now, research scientist Yuji Tsunekawa in the Laboratory for Cell Asymmetry (Fumio Matsuzaki, Team Leader), together with Keiichiro Suzuki and Juan Carlos Izpisua Belmonte at the Salk Institute of Biological Studies, USA, and other collaborators, has developed of a novel genome-editing technology, which exploits the NHEJ repair mechanism with the CRISPR/Cas9 system called, homology-independent targeted integration (HITI). This technology allows them to insert transgenes into both proliferating and non-proliferating cells efficiently. Their work, published in Nature, further demonstrates that HITI can be used for gene editing in vivo in the mouse and rat models.
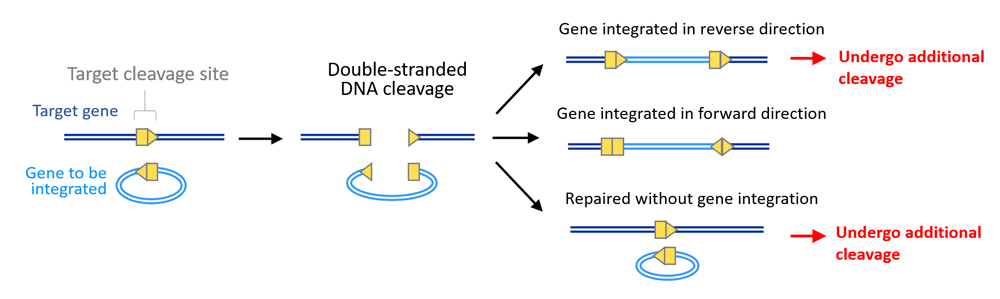
HITI donor vectors were constructed to ensure robust gene integration only when inserted in the forward direction, and if inserted in the reverse direction or is unintegrated, the DNA would undergo further cleavage by Cas9 until inserted correctly or gRNA is no longer able to bind to target sequences due to errors during NHEJ repair (see scheme above). When the HITI donor vector was tested in human cell lines, the team observed gene integration at target sites at efficiencies approximately ten times higher than conventional HDR-mediated methods. Effectiveness of HITI in non-proliferating cells was also examined using mouse primary neurons, in which they found gene insertion in 60% of transfected cells.
The team next tested whether their method can be used in non-dividing cells in vivo. The adeno-associated virus (AAV) expressing Cas9 systems and HITI donor was injected directly into different tissues of adult mice, such as the brain and muscles, and confirmed that transgenes were correctly knocked in some cells in the vicinity of injection sites. When HITI-AAV vectors were delivered intravenously just after birth, gene insertion was confirmed in a small percentage (3–10 %) of cells within various tissues of the body. The team also injected their HITI-AAV vector in the eye of a rat model for retinitis pigmentosa (RP), after which they detected a rise in mRNA expression levels of Merkt, a gene implicated in RP, as well as thickening of degenerating photoreceptor layer.
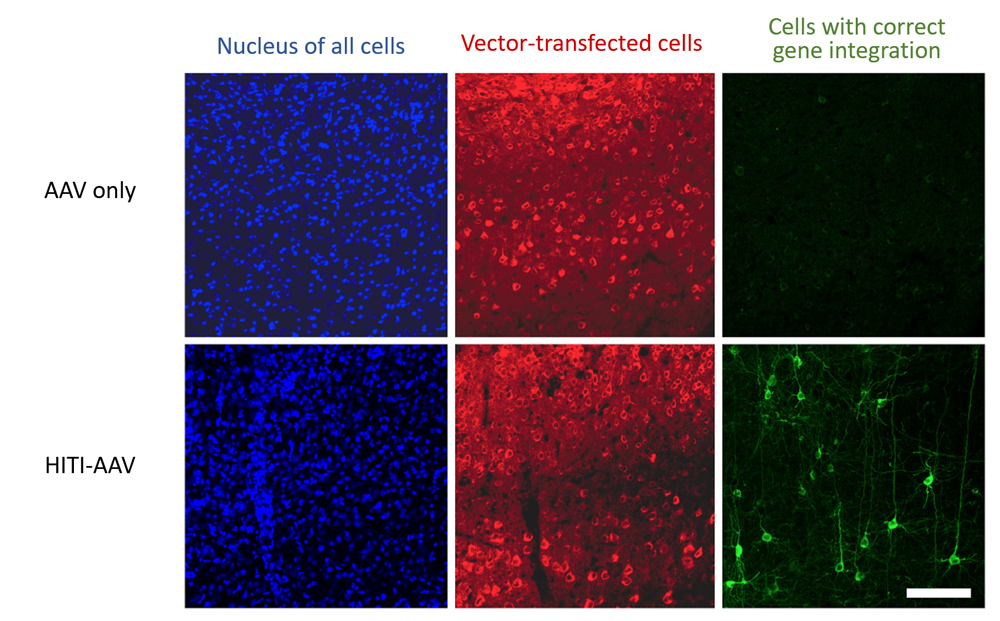
“As efficiency of gene insertion in vivo is still low and some mistargeting and mutations of target genes do occur, there is still much work that need to be done to improve the HITI method before it can be used in clinical applications. Nevertheless, our study clearly demonstrates that in vivo genome editing is possible,” explains Tsunekawa. “The HITI technology is sure to become a powerful tool not only for clinical applications, but also for basic research. It can be used to visualize and track fates of specific cells by inserting fluorescent marker genes, or modify genomes in animals, such as non-human primates, for which generation of transgenic models has been difficult.”
| Link to article |
In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. |
|---|